Abstract
Fagus multinervis is an endemic plant found in Ulleungdo Island in Korea. We present a second complete chloroplast genome of Fagus multinervis. The length of chloroplast genome is 158,349 bp, consisting of four subregions: 87,660 bp of LSC and 18,903 bp of SSC regions separated by a pair of 25,893 bp of IR regions. It includes 128 genes (83 protein-coding genes, eight rRNAs, and 37 tRNAs). There are two SNPs and two INDELs between two accessions of F. multinervis. The phylogenetic tree shows that the two accessions of F. multinervis form a clade, sister to F. engleriana.
Fagus multinervis Nakai is a tree and endemic to Ulleungdo Island in Korea, a small oceanic island originated about 1.8 million years ago (Kim Citation1985). It plays an important ecological role in the deciduous forest on the island (Oh Citation2015; Oh et al. Citation2016). High genetic diversity was shown based on allozyme (Chung et al. Citation1998; Ohkawa et al. Citation2006), but only a single chloroplast haplotype was found based on the psbA-trnH region (Oh Citation2015). To understand the chloroplast haplotype diversity and spatial distribution of F. multinervis within Ulleungdo, we determined the second complete chloroplast genome of the species.
Total genomic DNA was extracted from fresh leaves of F. multinervis collected from Ulleungdo Island (37°29′19.0″N, 130°53′15.9″E; voucher in the herbarium of Daejeon University (TUT); Oh 7738) by using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 at Macrogen Inc., and de novo assembly was done by the method described in Park J-S et al. (Citation2019). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for genome annotation based on previously reported F. multinervis chloroplast genome (MK518070).
The length of assembled chloroplast genome of F. multinervis in this study (GenBank accession number: MN894556) is 158,349 bp (GC ratio is 37.1%) with four subregions: 87,660 bp of large single-copy (35.0%), 18,903 bp of small single-copy (31.1%) regions, and 25,893 bp of a pair of inverted repeats (42.7%). It contains 128 genes (83 protein-coding genes, 8 rRNAs, and 37 tRNAs); 18 genes (seven protein-coding genes, four rRNAs and seven tRNAs) are duplicated in the IR regions.
Two single nucleotide polymorphisms (SNPs) and two insertions and deletions (INDELs) were identified from the alignment with the earlier chloroplast genome of F. multinervis (Park J-S et al. Citation2019), which will be useful resources to develop molecular markers to measure population structure of the species. One synonymous SNP is in psbM and one non-synonymous SNP is found in ccsA. The level of molecular variation of the species is large compared to other woody species in Korea. In Camellia japonica, one each of SNP and INDEL were found between Jejudo and Soyeonpyeongdo Islands in Korea (Park J, Kim Y, Xi H, et al. Citation2019). Only one SNP was found in Abeliophyllum distichum (Min et al. Citation2019), and no variation was reported in Salix koriyanagi (Park J, Kim Y, Xi H Citation2019). However, 265 SNPs and 247 INDELs are found between F. multinervis and F. engleriana, suggesting that the chloroplast genomes of the two species is highly divergent.
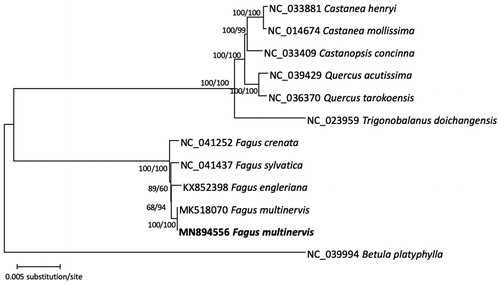
Eleven complete chloroplast genomes of Fagaceae including two F. multinervis were used for constructing neighbor-joining and maximum likelihood trees using MEGA X (Kumar et al. Citation2018) based on the alignment of complete chloroplast genomes by MAFFT 7.450 (Katoh and Standley Citation2013). The phylogenetic tree shows that Fagus is sister to all other Fagaceae, consistent with previous study (Oh and Manos Citation2008) and two accessions of F. multinervis forms a clade, sister to F. engleriana ().
Figure 1. Neighbor-joining and maximum-likelihood phylogenetic tree based on multiple sequence alignment of 11 Fagaceae chloroplast genomes: Fagus multinervis (MN894556 in this study and MK518070), F. engleriana (KX852398), F. sylvatica (NC_041437), F. crenata (NC_041252), Trigonobalanus doichangensis (NC_023959), Quercus tarokoensis (NC_036370), Q. acutissima (NC_039249), Castanopsis concinna (NC_033409), Castanea mollissima (NC_014674), and C. henryi (NC_033881). Betula platyphylla (NC_039994) was used as an outgroup. Phylogenetic tree was drawn on neighbor-joining tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum-likelihood phylogenetic trees.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chung H, Chung J, Chung M. 1998. Allozyme variation in six flowering plant species characterizing Ullung Island, Korea. J Jap Bot. 73:241–247.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mole Biol Evol. 30(4):772–780.
- Kim YK. 1985. Petrology of Ulreung Volcanic Island. J Japan Assoc Min Petr Econ Geol. 80(7):292–303.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Min J, Kim Y, Xi H, Jang T, Kim G, Park J, Park J-H. 2019. The complete chloroplast genome of a new candidate cultivar, Sang Jae, of Abeliophyllum distichum Nakai (Oleaceae): initial step of A. distichum intraspecies variations atlas. Mitochondrial DNA Part B. 4(2):3716–3718.
- Oh S-H. 2015. Sea, wind, or bird: origin of Fagus multinervis (Fagaceae) inferred from chloroplast DNA sequences. Korean J Pl Taxon. 45(3):213–220.
- Oh S-H, Manos PS. 2008. Molecular phylogenetics and cupule evolution in Fagaceae as inferred from nuclear CRABS CLAW sequences. Taxon. 57:434–451.
- Oh S-H, Youm J-W, Kim Y-I, Kim Y-D. 2016. Phylogeny and evolution of endemic species on Ulleungdo island, Korea: the case of Fagus multinervis (Fagaceae). Syst Bot. 41(3):617–625.
- Ohkawa T, Kitamura K, Takasu H, Kawano S. 2006. Genetic variation in Fagus multinervis Nakai (Fagaceae), a beech species endemic to Ullung Island, South Korea. Plant Species Biol. 21(3):135–145.
- Park J-S, Jin D-P, Park J-W, Choi B-H. 2019. Complete chloroplast genome of Fagus multinervis, a beech species endemic to Ulleung Island in South Korea. Mitochondrial DNA Part B. 4(1):1698–1699.
- Park J, Kim Y, Xi H. 2019. The complete chloroplast genome sequence of male individual of Korean endemic willow, Salix koriyanagi Kimura (Salicaceae). Mitochondrial DNA Part B. 4(1):1619–1621.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019. The complete chloroplast genome of common camellia tree in Jeju island, Korea, Camellia japonica L. (Theaceae): intraspecies variations on common camellia chloroplast genomes. Mitochondrial DNA Part B. 4(1):1292–1293.
