Abstract
Dendrobium pseudotenellum is an endangered species in Orchidaceae, which is distributed in Yunnan of China and Vietnam. Here, we report the complete chloroplast (cp) genome sequence and its genome features of D. pseudotenellum. The cp genome sequence of D. pseudotenellum was 149,114 bp in length with 37.4% GC content. It presented a typical quadripartite structure including a large single-copy region (LSC, 84,262 bp), a small single-copy region (SSC,12,786 bp), and two inverted repeat regions (IRA and IRB, 26,033 bp, each). The cp genome encoded 125 genes, including 80 protein-coding genes, 37 tRNAs, and 8 rRNAs. The phylogenetic analysis was conducted based on 72 protein-encoding genes from D. pseudotenellum and other 19 Dendrobium species, with 4 species of Cymbidium and Pleione as outgroup. It turned out that D. pseudotenellum was a sister to D. exile. This chloroplast genome would be helpful for the phylogeny and conservation of Dendrobium.
Dendrobium pseudotenellum Guillaum is a perennial epiphytic orchid in mountain forests and distributed in China (S. Yunnan) to Vietnam. It was characterized by subterete leaves with small and white flowers, which is placed in the Sect. Crumenata Pfitz (Chen et al. Citation2009). Owing to its narrow distribution and highly medicinal value, D. pseudotenellum was listed as an endangered species in the China Species Red List (Wang and Xie Citation2004; Wang et al. Citation2020). Here, we report the cp genome of D. pseudotenellum in order to add molecular data on the phylogeny and conservation of Dendrobium.
Leaf samples of D. pseudotenellum were collected from Fumin County, Yunnan Province, China (25°20′19ʺN, 102°27′26ʺE). The voucher specimen was deposited in the Herbarium of Southwest Forestry University (HSFU, specimen code of Lilu20180003). The cp genomic DNA was extracted from the fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987), and sequenced by Illumina Hiseq 2500 platform. Genome sequences were screened out and assembled with Geneious Prime version 2020.0.4 (Kearse et al. Citation2012), with the chloroplast genome of D. officinale Kimura et Migo (GenBank Accession No. NC 024019) as the reference sequences. Finally, we submitted the sequence data to GenBank with an officially accession number MN 915016.
The complete chloroplast genome of D. pseudotenellum was 149,114 bp in length, which presented a typical quadripartite structure, containing a large single-copy region (LSC 84,262 bp), a small single-copy region (SSC 12,786 bp), and two inverted repeat regions (IRA and IRB, 26,033 bp). Besides, the complete chloroplast genome of D. pseudotenellum encoded 125 genes, including 80 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. In addition, the overall GC content of the whole plastome was 37.4%, whereas the corresponding values of the LSC, SSC, and IR regions were 34.9%, 30.2%, and 43.3%, respectively. The gene order and organization of the D. pseudotenellum were consistent with those of other species in Dendrobium (Huang et al. Citation2019; Wang et al. Citation2020).
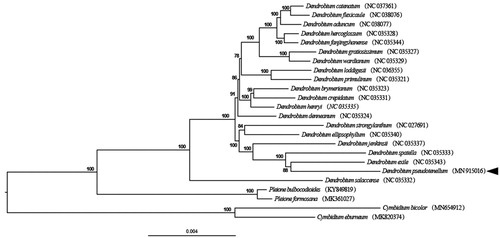
To investigate the position of D. pseudotenellum, a maximum-likelihood (ML) phylogenetic analysis was performed based on 72 protein-coding genes from 20 species in Dendrobium, with 4 species in Pleione D. Don. and Cymbidium Sw. as outgroup. All the data were downloaded from NCBI GenBank. All sequences were aligned with the Geneious Prime version 2020.0.4. Then, the phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) with 1000 ultrafast bootstrap (UFBoot) replicates (Minh et al. Citation2013). Results revealed that D. pseudotenellum was a sister to D. exile with 88% bootstrap values ().
Acknowledgements
We are grateful to Dr. Fei Zhao in Kunming Institute of Botany Chinese Academy of Sciences, for his help in preparing this manuscript. We also thank Mr. Zhi-Feng Xu and Mrs. Xiao-Yun Wang in the Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Co., Ltd, for providing sample materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Ji ZH, Luo YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 25. St. Louis (MO): Missouri Botanical Garden Press; p. 1–506.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang ZC, Pan YY, Chen GZ, Chen LJ, Wu XY, Huang J. 2019. The complete chloroplast genome of Dendrobium harveyanum (Orchidaceae). Mitochondrial DNA Part B. 4(2):3200–3201.
- Kearse M, Moir R, Wilson A, Havas SS, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang S, Xie Y. 2004. China species red list. RedList. Vol. 1. Beijing (China): Higher Education Press.
- Wang YP, Ai J, Luo Y, Li QQ, Li L. 2020. The complete chloroplast genome of Dendrobium wattii (Orchidaceae. ). Mitochondrial DNA Part B. 5(2):1325–1326.