Abstract
Dendrobium chrysocrepis is an endangered and rare species belonging to the family Orchidaceae. Here, we report the complete chloroplast (cp) genome sequence and the cp genome features of D. chrysocrepis. The complete cp genome sequence of D. chrysocrepis was 154,155 bp in length and presented a typical quadripartite structure including one large single-copy region (LSC, 86,372 bp), one small single-copy region (SSC, 15,463 bp), and two inverted repeat regions (IR, 26,160 bp). The cp genome encoded 130 genes, of which 115 were unique genes. The phylogenetic relationships indicated that D. chrysocrepis was closely related to other species in Dendrobium.
The genus Dendrobium Sw. is one of the largest genera in the family Orchidaceae with about 1000 species, which are mainly distributed in tropics and subtropics in Asia, Oceanica, and Pacific islands (Pridgeon et al. Citation2014; Teixeira da Silva et al. Citation2016). Dendrobium chrysocrepis C. S. P. Parish & Rchb. f. ex Hook. f. is a very rare, special, and endangered species. It is similar to D. moschatum (Buch.-Ham.) Sw., but can be easily distinguished by having solitary flowers with spoon-like lip which are borne on the flat compressed stem (Jin and Li Citation2006; Phueakkhlai et al. Citation2018). The last records came from Thailand (Phueakkhlai et al. Citation2018) and China (Yunnan) (Jin and Li Citation2006; Li et al. Citation2009). Considering that cp genome played an important role in revealing the origin of the evolution and phylogeny of Dendrobium (Pan et al. Citation2019; Wang et al. Citation2020), the complete cp genome of D. chrysocrepis was assembled in this paper.
Leaf samples of D. chrysocrepis were obtained from Fumin County (25°20′19ʺN, 102°27′26ʺE), Yunnan Province, China. The specimen was deposited in the Herbarium of Southwest Forestry University (HSFU, Lilu20180001). Total genomic DNA was extracted from fresh leaves using the modified CTAB procedure (Doyle and Doyle Citation1987). It was sequenced on Illumina Hiseq 2500 platform (Illumina, San Diego, CA). With the chloroplast genome of D. officinale Kimura et Migo (GenBank accession number NC024019) as the reference sequences, we assembled the complete chloroplast genome from the clean reads by the GetOrganelle pipe-line (Jin et al. Citation2018), and then annotated the new sequences using the Geneious Prime version 2020.0.4 (Kearse et al. Citation2012). Finally, the complete cp genome sequence was submitted to the GenBank with accession number MN915015.
The complete cp genome sequence of D. chrysocrepis was 154,155 bp in length and consists of a large (LSC, 86,372 bp) and a small (SSC, 15,463bp) single-copy regions, separated by a pair of identical inverted repeats (IR, 26,160 bp). The cp genome encoded 130 genes, of which 115 were unique genes (77 protein-coding genes, 30 tRNAs, and 8 rRNAs). The overall GC content was 37.5%, whereas the corresponding values of the LSC, SSC, and IR regions were 35.2%, 30.6%, and 43.5%, respectively.
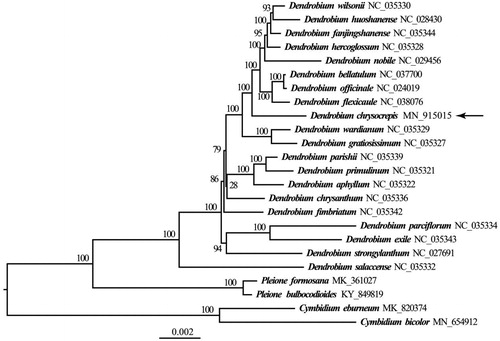
To confirm the phylogenetic position of D. chrysocrepis, a molecular phylogenetic tree was constructed based on 72 protein-coding genes from 20 Dendrobium species, with 4 species from Cymbidium Sw. and Pleione D. Don as outgroup. The maximum-likelihood (ML) analysis was performed using the CIPRES Science Gateway web server (RAxML-HPC2 on XSEDE 8.2.10) with 1000 bootstrap replicates and settings as described by Stamatakis et al. (Citation2008). The results showed that D. chrysocrepis was closely related to other species in Dendrobium (). This new data on the cp genome of D. chrysocrepis provided an essential clue for better understanding of the phylogeney and biodiversity of Dendrobium.
Acknowledgements
We are grateful to Dr. Fei Zhao in Kunming Institute of Botany Chinese Academy of Sciences, for his help in preparing this manuscript. We also thank Mr. Zhi-Feng Xu and Mrs. Xiao-Yun Wang in the Wild Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Company, for providing sample materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv: 256479.
- Jin XH, Li H. 2006. Coelogynetsii and Dendrobium menglaensis (Orchidaceae), two new species from Yunnan, China. Ann Bot Fennici. 43:295–297.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li L, Ye DP, Li JW, Xing FW. 2009. A newly recorded species and a new synonym of Orchidaceae from China. J Trop Subtrop Bot. 17(3):295–297.
- Pan Y-Y, Li T-Z, Chen J-B, Huang J, Rao W-H. 2019. Complete chloroplast genome of Dendrobiumthyrsiflorum (Orchidaceae). Mitochondrial DNA Part B. 4(2):3192–3193.
- Phueakkhlai O, Suddee S, Hodkinson TR, Pedersen HAE, Srisom P, Sungkaew S. 2018. Dendrobium chrysocrepis (Orchidaceae), a new record for Thailand. Thai Forest Bull Bot. 46:134–137.
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. 2014. Genera Orchidacearum 6. Epidendroideae (part three). Oxford: Oxford University Press.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 57(5):758–771.
- Teixeira da Silva JA, Jin XH, Dobránszki J, Lu JJ, Wang HZ, Zotz G, Cardoso JC, Zeng SJ. 2016. Advances in Dendrobium molecular research: applications in genetic variation, identification and breeding. Mol Phylogenet Evol. 95:196–216.
- Wang YP, Ai J, Luo Y, Li QQ, Li L. 2020. The complete chloroplast genome of Dendrobium wattii (Orchidaceae). Mitochondrial DNA Part B. 5(2):1325–1326.