Abstract
Epimedium rhizomatosum is a rare endemic plant species inhabited in some high-elevation mountain area in the Sichuan province of China. In this study, we assembled the complete chloroplast genome of E. rhizomatosum from Illumina short-read sequencing data. The chloroplast genome of E. rhizomatosum was 159,151 bp in length, which comprises of a large single copy (LSC) region of 86,604 bp, a small single copy (SSC) region of 17,071 bp and two inverted repeat regions (IRa and IRb) of 27,737 bp for each. The total GC content of E. rhizomatosum chloroplast genome was 38.8%, while the corresponding values of LSC, SSC, and IR regions were 37.3%, 32.8%, and 43.0%, respectively. A total of 112 unique genes was identified from the chloroplast genome of E. rhizomatosum, including 78 protein-coding genes, four ribosomal RNA genes, and 30 tRNA genes. The phylogenetic analysis showed that E. rhizomatosum closely related to E. acuminatum. Our study will provide useful information on further clarifying the phylogenetic and evolutionary relationship in the genus Epimedium.
Epimedium L. is an herbaceous genus in berberidaceae that belongs to the basal eudicot plant family. Epimedii folium is a well-known Traditional Chinese medicine in China, which has been used to treat sexually-related dysfunction since two thousands year ago in China. In recent years, it was proven that Epimedii herba extract (EE) had therapeutic effects on liver cancer, osteoporosis, and menonpausal syndrome (Jiang et al. Citation2015; Zhang et al. Citation2016).
There are more than 58 species in the genus Epimedium, most of them closely distributed in the southwest of China. The phylogenetic relationship of some species in this genus remain unclear due to lack of efficient research approach. Recent studies showed that the complete chloroplast genome sequence as a super-barcode is a potent tool to assess the taxonomic and phylogenetic relationship. So far, just some species in the genus Epimedium have been reported (Liu et al. Citation2019; Zhang et al. Citation2016), there is still more species to be sequenced and assembled.
E. rhizomatosum is a rare endemic plant species in some high-elevation mountain area in the Sichuan province of China. The taxonomic status of E. rhizomatosum is controversial. In this study, we assembled the complete chloroplast genome of E. rhizomatosum from Illumina short-read sequencing data. The phylogenetic analysis showed that E. rhizomatosum closely related to E. acuminatum. Our study will provide useful information on further clarifying the phylogenetic and evolutionary relationship in the genus Epimedium.
In present study, a wild individual of E. rhizomatosum was collected from the Leibo County of Sichuan Province, China (N28°16′, E103°34′). The voucher sample (K201901) was deposited in the Herbarium of the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences, Beijing, China. The genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle 1987). A 300-bp shotgun library was prepared from the total genomic DNA. The library was sequenced on an Illumina Novaseq PE150 platform. The generated 150 bp paired-end reads were cleaned by removing adapter and low quality reads. The complete chloroplast genome was assembled from clean reads using the assembly pipeline of GetOrganelle v1.5 (Jin et al. Citation2018) that integrates the assembler SPAdes v3.9.0 (Bankevich et al. Citation2012). The chloroplast genome was annotated by the online program GeSeq (Tillich et al. Citation2017) and CPGAVAS2 (Shi et al. Citation2019), followed by careful manual correction. The annotated chloroplast genome of E. rhizomatosum has been deposited in GenBank with an accession number MN867588.
The chloroplast genome of E. rhizomatosum is 159,150 bp in length, which comprises of a large single copy (LSC) region of 86,605 bp, a small single copy (SSC) region of 17,071 bp and two inverted repeat regions (IRa and IRb) of 27,737 bp for each. The total GC content of E. rhizomatosum chloroplast genome is 38.8%, whereas the corresponding values of LSC, SSC, and IR regions are 37.3%, 32.8%, and 43.0%, respectively.
A total of 112 unique genes was identified from the chloroplast genome of E. rhizomatosum, including 78 protein-coding genes, four ribosomal RNA genes, and 30 tRNA genes. The majority of these genes are present in single copy, but 17 genes have two copies. A total of 18 genes were found to have introns in the chloroplast genome of E. rhizomatosum. Among these genes, atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC have one intron, whereas clpP, rps12, and ycf3 contains two introns.
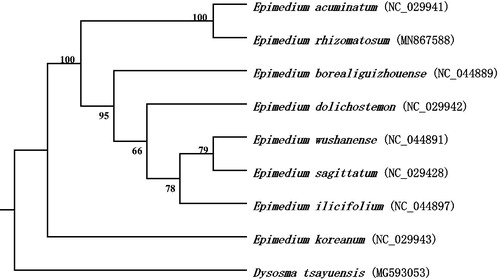
To analyze the phylogenetic relationship of E. rhizomatosum, we downloaded the complete chloroplast genome sequences of seven Epimedium species from the NCBI GenBank database. The phylogenetic tree was generated based on whole chloroplast genome sequences. MAFFT v7 (Katoh et al. Citation2017) was used to align the genome sequences, and then a maximum likelihood tree was constructed by using the software of RAxML v8.2.10 (Stamatakis Citation2014), with Dysosma tsayuensis as the outgroup (). The phylogenetic analysis showed that E. rhizomatosum closely related to E. acuminatum. Our study will provide useful information on further clarifying the phylogenetic and evolutionary relationship in the genus Epimedium.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jiang J, Song J, Jia X. 2015. Phytochemistry and ethnopharmacology of Epimedium L. Species. Chin Herb Med. 7(3):204–222.
- Jin J, Yu W, Yang J, Song Y, Yi T, Li D. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 4:1–7.
- Liu X, Yang Q, Zhang C, Shen G, Guo B. 2019. The complete chloroplast genome of Epimedium sagittatum (Sieb. Et Zucc.) Maxim. (Berberidaceae), a traditional Chinese herb. Mitochondrial DNA Part B. 4(2):2572–2573.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee S, Yang T, et al. 2016. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 7:306