Abstract
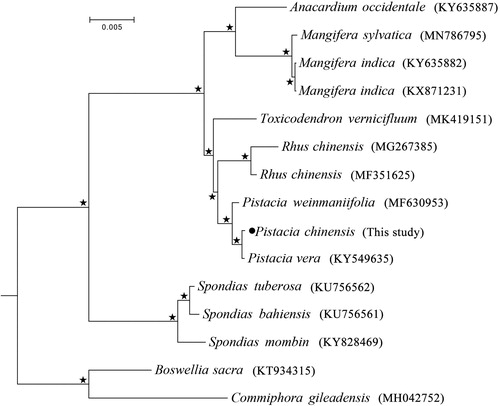
Pistacia chinensis Bunge (Anacardiaceae: Rhoideae) is a rare resource plant that originated in China, and has been widely used in horticultural and production field. In this study, we sequenced the complete chloroplast genome of P. chinensis on the Illumina platform by shotgun genome skimming method. The complete chloroplast genome of P. chinensis was 160,596 bp in length with the GC content 37.9%, which was comprised of a large single copy (LSC) region of 88,310 bp, a small single copy (SSC) region of 19,090bp, and a pair of inverted repeats (IRA/IRB) of 26,598 bp. The chloroplast genome encoded 132 genes including 87 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Phylogenetic analysis constructed using maximum likelihood (ML) method strongly supported the monophyly of each genus used in the family Anacardiaceae, and the genus Pistacia is a sister group to the genus Rhus with 100% bootstrap value. Pistacia chinensis Bunge was sister to P. vera and then grouped with Pistacia weinmaniifolia as a cluster.
Pistacia chinensis Bunge (Anacardiaceae: Rhoideae), a small to medium-sized tree, can grow up in harsh environment and poor soils (Min and Barfod Citation2012). As a green energy, the oil from the seeds has been considered a promising option for biodiesel, and gained more and more attention (Fu et al. Citation2009). Also, the species is popularly used in horticultural design, and production of furniture and yellow dye (Min and Barfod Citation2012). Pistacia chinensis is native to China, and widely distributed with records in 23 provinces (Wang Citation2005). There was only one report regarding the complete chloroplast genome of P. chinensis until now (Xu et al. Citation2019). Here, we used the shotgun genome skimming method to sequence the complete chloroplast genome of P. chinensis in order to better understand its genome structure and organization, and analyze its phylogenetic relationship with other Anacardiaceae species to provide more data for its original and evolutionary research.
We collected the samples of Pistacia chinensis in Bazhong, Sichuan, China (31°9′36″N, 106°12′36″E) in July, 2014. The specimen is stored at the Herbarium of School of Life Science, Shanxi University, China (voucher no Ren_P1382). We extracted the genomic DNA from the leaves and obtained the chloroplast sequence of P. chinensis by the shotgun genome-skimming method on an Illumina HiSeq 4000 platform (Zimmer and Wen Citation2015). The complete chloroplast genome was assembled using GetOrganelle program (Jin et al. Citation2018) and annotated using DOGMA (Wyman et al. Citation2004) and Geneious (version 11.0.3; https://www.geneious.com).
The complete chloroplast genome of Pistacia chinensis was 160,596 bp in length (GenBank Accession No. MT157378), and consisted four distinct parts which included a large single copy (LSC) region of 88,310 bp, a small single copy (SSC) region of 19,090 bp, and a pair of inverted repeats (IRA/IRB) of 26,598 bp. The LSC region contained 60 protein-coding and 22 tRNA genes, whereas the SSC region contained 11 protein-coding and one tRNA genes. Eight protein-coding genes and 7 tRNAs were duplicated in the IR region. All rRNA genes were located in the IR region. The chloroplast genome totally encoded 132 genes including 87 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The overall nucleotide composition is 30.8% A, 19.3% C, 18.6% G and 31.4% T with the total GC content 37.9%.
We downloaded the chloroplast genome of 14 Anacardiaceae species from GenBank with the two species Commiphora gileadensis and Boswellia sacra from the family Burseraceae as outgroups. Based on all the chloroplast protein-coding genes, we performed the phylogenetic analysis using RAxML program under GTRGAMMA model with 1000 bootstrap replicates (Stamatakis Citation2014). The results () strongly supported the monophyly of each genus used in the family Anacardiaceae, and the genus Pistacia is a sister group to the genus Rhus with well-support (100% BS value). Pistacia chinensis Bunge was sister to P. vera and then grouped with Pistacia weinmaniifolia as a cluster. The complete chloroplast genome of Pistacia chinensis would provide valuable genetic information for the further conservation and evolutionary researches.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Fu Y, Pan XB, Gao H. 2009. Geographical distribution and climate characteristics of habitat of Pistacia chinensis Bunge in China. Chinese J of Agr. 30(3):318–322.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. DOI:10.1101/256479.
- Min TL, Barfod A. 2012. Pistacia chinensis Bunge, Enum. Pl. China Bor. 15. 1833”. Flora of China. Missouri Botanical Garden, St. Louis, MO and Harvard University Herbaria, Cambridge, MA. [Retrieved Sep 14].
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang T. 2005. A survey of the woody plant resources for biomass fuel oil in china. Science Rev. 23(5):12–14.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xu JH, Zhang DX, Sun K, Wang XR, Xiang QH, Wang Q, Guan WB. 2019. The complete chloroplast genome sequences of Pistacia chinensis Bunge, a potential bioenergy tree. Mitochondrial DNA Part B. 4(1):1774–1775.
- Zimmer EA, Wen J. 2015. Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. J Syst Evol. 53:371–379.