Abstract
Ricania speculum is called as black planthopper, belonging to the family Ricaniidae. We have determined a mitochondrial genome of fungal endosymbiont of R. speculum, belonging to the family Ophicocordycipitaceae. The circular mitogenome of this fungus is 66,785 bp including 32 protein-coding genes, 2 ribosomal RNA genes, and 19 transfer RNAs. Its GC ratio is 30.6%. A phylogenetic tree presented that this fungus is clustered with fungi of two genera, Hirsutella and Ophiocordyceps.
Ricania speculum (Walker, 1851) is called as black planthopper belonging to the family Ricaniidae of the order Hemiptera. It has been known that insect species in this family contains fungal symbionts or pathogens (Mueller Citation2011). For example, Scolypopa australis has entomopathogen, Lecanicillium muscarium (Marshall et al. Citation2003) and Ricania japonica contains yeast-like symbionts (Hongoh and Ishikawa Citation2000). When we conducted de novo assembly using NGS raw reads obtained from a R. speculum sample collected in Jungtosa Template in Jeollabuk-do Province in Korea (35.6585365 N, 126.887853E; the specimen is stored in Gyeongsang National University, Korea, Accession number: 2018-HJ-56), a circular fungal mitogenome sequence was obtained together with a R. speculum mitogenome. There is no sequence variation on this fungal mitochondrial genome based on the alignment of raw reads, reflecting that the amount of this fungus inside R. speculum occupies relatively large and homogeneous group; it can be a fungal symbiont of R. speculum.
DNA was extracted using DNeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina HiSeq2000 (Macrogen Inc., Korea) were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008) and gaps were closed with SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17, and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitochondrial genome with referring mitogenomes of Tolypocladium inflatum (NC_036382; Zhang et al. Citation2017) and Fusarium oxysporum (MN259514; Kwon et al. Citation2019).
The mitogenome of fungal symbiont of R. speculum (MT019333) is 66,785 bp long and its GC ratio is 30.3%. It contains 32 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), and 19 transfer RNA genes (tRNAs). Number of tRNAs in this mitochondrial genome is smaller than the other fungal mitochondrial genomes missing three types of tRNAs: tRNA-Ala, tRNA-Gln, and tRNA-Tyr. Thirteen out of 32 PCGs are identified as endonuclease, which is one of the contributors to expand Aspergillus and Penicillium mitochondrial genomes (Joardar et al. Citation2012; Park et al. Citation2019).
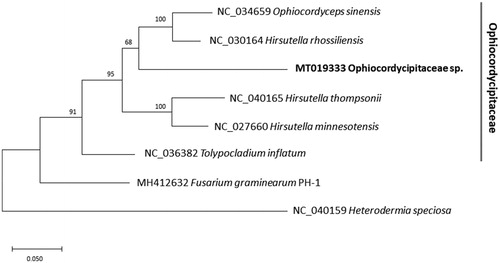
We inferred the phylogenetic relationship of six representative mitogenomes belonging to the family Ophiocordycipitaceae with Fusarium graminearum (Nectriaceae) and Heterodermia speciosa (Physciaceae) as outgroup species. Multiple sequence alignment was conducted by MAFFT 7.450 (Katoh and Standley Citation2013) based on concatenated the 12 conserved genes selected based on annotation of the eight mitochondrial genomes. A maximum likelihood phylogenetic tree was constructed using MEGA X (Kumar et al. Citation2018). Phylogenetic trees present that our species is clustered with Hirsutella and Ophiocordyceps spp. (; Simmons et al. Citation2015). While rRNA of this mitogenome displays that 85.94% identity against Tolypocladium inflatum as a best hit, which is incongruent with phylogenetic tree. This mitogenome is one of the examples which shows power of NGS technologies to understand unknown fungal symbionts in insect (e.g. aphid mitogenome from Nymphaea colodata genome (Park et al. accepted) and Stegobium paniceum mitogenome from the truffle (Tuber melanosporum) whole genomes (Park et al. in preparation) as well as contributing fungal mitogenome sequence resources for further diverse researches.
Figure 1. Maximum likelihood (1,000 bootstrap repeats) phylogenetic tree of 12 conserved genes, ATP6, ATP8, COB, COX1, COX2, COX3, NAD1, NAD3, NAD4, NAD4L, NAD5, and NAD6, originated from the six mitochondrial genomes of Ophiocordycipitaceae and two outgroup species: Ophiocordycipitaceae sp., Ophiocordyceps sinensis, Hirsutella rhossiliensis, Hirsutella thompsonii, Hirsutella minnesotensis, and Tolypocladium inflatum and Fusarium graminearum PH-1and Heterodermia speciose as two outgroup species. Gray line indicates family of six fungal species used in the tree. The numbers above branches indicate bootstrap support values of a maximum likelihood phylogenetic tree.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Hongoh Y, Ishikawa H. 2000. Evolutionary studies on uricases of fungal endosymbionts of aphids and planthoppers. J Mol Evol. 51(3):265–277.
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genom. 13(1):698.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwon W, Park J, Kim J-B, Park M-J, Kim T-S. 2019. Complete mitochondrial genome sequence of lettuce pathogenic fungus, Fusarium oxysporum f. sp. lactucae 16-086. Mitochondrial DNA Part B. 4(2):3227–3228.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Marshall R, Lester M, Glare T, Christeller J. 2003. The fungus, Lecanicillium muscarium, is an entomopathogen of passionvine hopper (Scolypopa australis). NZ J Crop Hortic Sci. 31(1):1–7.
- Mueller GM. 2011. Biodiversity of fungi: inventory and monitoring methods. Burlington, MA: Elsevier, Academic Press.
- Park J, Kwon W, Huang X, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of a xerophilic fungus Aspergillus pseudoglaucus. Mitochondrial DNA Part B. 4(2):2422–2423.
- Simmons DR, Kepler RM, Renner SA, Groden E. 2015. Phylogeny of Hirsutella species (Ophiocordycipitaceae) from the USA: remedying the paucity of Hirsutella sequence data. IMA Fungus. 6(2):345–356.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang Y-J, Yang X-Q, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101(23-24):8517–8531.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
