Abstract
The complete chloroplast genome sequence of Swertia souliei, an alpine annual herbaceous plant endemic to the Qinghai-Tibetan Plateau (QTP), is reported in this study. The plastome is 152,804 bp in length, with one large single-copy region of 83,195 bp, one small single-copy region of 18,096 bp, and two inverted repeat (IR) regions of 25,756 bp. It contains 130 genes, including 85 protein-coding, 8 ribosomal RNA, and 37 transfer RNA genes. Phylogenetic tree shows that this species is a sister to the clade containing S. leducii, S. verticillifolia, and S. mussotii. The complete chloroplast genome could provide significant insight for understanding the phylogenetic relationship of taxa within Gentianaceae.
Swertia souliei Burkill, belonging to genus Swertia (Gentianaceae), is an alpine annual herbaceous plant endemic to the Qinghai-Tibetan Plateau (QTP) (Ho and Liu Citation2001). It is mainly distributed in Sichuan Provinces, occurring primarily in alpine meadow, with an elevation of 3700–4400 m.a.s.l. (Ho and Liu Citation2001). As a kind of traditional Chinese medicine, this species is sufferring over exploitation and reduced natural habitats and needs urgent conservation (Xiao et al., Citation2010). Knowledge of the genetic information of this species would contribute to the formulation of protection strategy. As one of the important targets for genetic transformation, the full chloroplast genome could supply more genetic information.
Fresh leaves of S. souliei were collected from Hongyuan (Sichuan, China; coordinates: 102°26′E, 30°20′N) and dried with the silica gel. The voucher specimen was stored in Sichuan University Herbarium with the accession number of QTP2017009. Total DNA was isolated using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced by the BGISEQ-500 sequencing platform (BIG, Shenzhen, China). A total of 10 million high-quality pair-end reads were used to assemble the complete chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017). The complete chloroplast genome sequence of S. leducii was used as a reference. Plann v1.1 (Huang and CronK Citation2015) and Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the chloroplast genome and correct the annotation.
The total plastome length of S. souliei (MT185926) is 152,804 bp and the plastome exhibits a typical quadripartite structural organization, consisting of a large single-copy (LSC) region of 83,195 bp, two inverted repeat (IR) regions of 25,756 bp, and a small single-copy (SSC) region of 18,096 bp. The chloroplast genome contains 130 complete genes, including 85 protein-coding genes (85 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 tRNA genes (37 tRNAs). Most genes occur in one single copy, while 15 genes occur in double, including 7 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) and 8 PCGs (rps7, rps12, rps19, rpl2, rpl23, ndhB, ycf15, and ycf2) with one partial rps19 gene being identified as a pseudogene. The overall AT content of the plastome is 61.9%, the corresponding values for the LSC, SSC, and IR regions are 63.8%, 68.1%, and 56.7%, respectively.
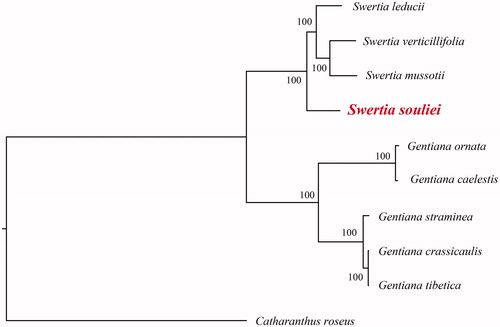
In order to further clarify the phylogenetic position of S. souliei, plastome of nine representative Gentianaceae species were obtained from NCBI to construct the plastome phylogeny, with Catharanthus roseus as an outgroup. All the sequences were aligned using MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood phylogenetic analyses were conducted using RAxML v.8.2.11 (Stamatakis Citation2014). The phylogenetic tree shows that all species were identified with two clades. G. ornata, G. caelestis, G. straminea, G. crassicaulis and G. tibetica clustered in one clade and the remaining Swertia species clustered in another clade (). The complete plastome sequence of S. souliei provides intragenic information for its conservation and contributes to research on the phylogenetic analyses of Gentianaceae.
Figure 1. A plastome phylogenetic tree of Gentianaceae species. GenBank accession numbers: Swertia leducii (NC_045301); Swertia verticillifolia (MF795137); Swertia mussotii (NC_031155); Gentiana ornata (NC_037983); Gentiana caelestis (MG192304); Gentiana straminea (KJ657732); Gentiana crassicaulis (NC_027442); Gentiana tibetica (NC_030319); Catharanthus roseus (NC_021423).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Ho TN, Liu SW. 2001. A worldwide monograph of Gentiana. Beijing: Science Press.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xiao YC, Wei LX, Yang HX, et al. 2010. Quality control of traditional tibetan medicine Swertia chirayita. J Chin Pharm Sci. 45:255–258.
