Abstract
The subalpine swordtail butterfly Graphium (Pazala) parus (Nicéville, 1900) is endemic to the Hengduan Mountains centered in West China, its biological and ecological characters still remain poorly understood. The present study reported the complete mitochondrial genome of this butterfly for the first time, which is the second mitogenome record for subgenus Pazala Moore, 1888. The mitogenome of G. (P.) parus is circular and 15,231 bp in length, and consists of 37 genes, including 13 PCGs, 22 tRNAs, and two rRNAs. The maximal likelihood (ML) tree containing the focal species and 30 other available Papilioninae members placed G. (P.) parus sister to G. (P.) mullah (Alphéraky, 1897) inside tribe Leptocircini, which agrees with the taxonomic characters of this species. The findings of this study added data to the complex genus Pazala and would benefit future evolution and conservation biology investigations.
The swordtail butterflies of the subgenus Pazala Moore, 1888 (Lepidoptera: Papilionidae) are a complex group of Sino-Himalayan species with morphological resemblance and geographical endemicity (Racheli and Cotton Citation2009); cryptic new taxa have been reported in very recent years (Hu et al. Citation2018; Citation2019). Graphium (Pazala) parus (Nicéville, 1900) is a narrow-ranged species endemic to the Hengduan Mountains of northwestern Yunnan, southeastern Tibet, and western Sichuan of China; as well as the very eastern margin of Kachin State, Myanmar. G. (P.) parus occupies the highest elevation range among all species in genus Graphium, and its biological and ecological characters are still poorly understood (Zhang et al. Citation2018). Since the mitochondrial genome harbors information related to adaptive evolution and conservation biology, the present study reports the complete mitogenome of G. (P.) parus, which is the second mitogenome published in subgenus Pazala, following the low-altitude species G. (P.) mullah (Alphéraky, 1897) (as Pazala timur) (Chen et al. Citation2014a).
The sample used in this study was collected from Sancha He (100.237254°E, 27.125564°N, 2,930 m) of Yulong Xueshan, Lijiang, Yunnan, China. The specimen was deposited in the zoological museum (insect collection) of Yunnan University, Kunming, China (specimen number: YNU-LEP-PAP-2018015). Genomic DNA was extracted from the thoracic muscle of a single male adult using phenol-chloroform and isopropanol protocol (Hu et al. Citation2013) and sequenced on an ABI 3730xl automatic sequencer (Applied Biosystems, CA, USA). Resultant gene fragments were assembled using DNAStar (https://www.dnastar.com/) with G. (P.) mullah (KJ472924) as the reference genome. Protein-coding genes (PCGs), transfer RNA genes (tRNAs) and ribosomal RNA genes (rRNAs) were predicted using the web based MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013).
The complete mitogenome of G. (P.) parus is circular and 15,231 bp in length (GenBank accession number: MT198821). The base composition is 40.09% for A, 39.48% for T, 7.85% for G, and 12.22% for C. This mitogenome contains 37 genes, including 13 PCGs, 22 tRNAs, and two rRNAs, plus a non-coding control region. The plus (+) strand encodes nine PCGs (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6, and cob), while the minus (−) strand encodes four PCGs (nad5, nad4, nad4l, and nad1). The gene arrangement and character of this genome fit those of Lepidoptera mitogenomes (Kim et al. Citation2009; Park et al. Citation2012).
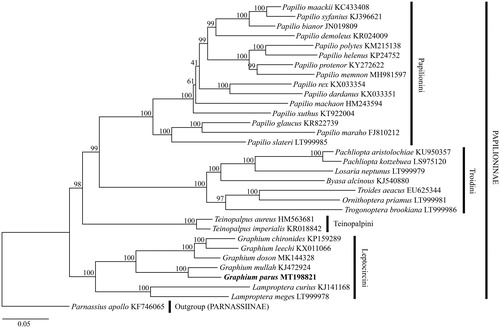
To validate the identity of this mitogenome, a maximum likelihood (ML) phylogenetic tree was reconstructed in MEGA 7.0 with the GTR + I + G model, and a bootstrap analysis with 10,000 replications was employed to test the robustness of the tree (Kumar et al. Citation2016). Thirty species of Papilioninae with available mitogenomes were used as ingroups in the tree analysis and Parnassius apollo Linnaeus, 1758 (Parnassiinae; KF746065) (Chen et al. Citation2014b) was chosen as the outgroup. The result shows that G. (P.) parus is sister to G. (P.) mullah and then other Graphium species within Leptocircini. All Graphium species used in this analysis form a monophyletic clade, supported by the maximal bootstrap values ().
Acknowledgments
We thank the Yulong Xueshan Provincial Nature Reserve Administration for assistance on the collection of specimens, and Adam M. Cotton (Chiang Mai, Thailand) for improving the earlier drafts of this article.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
The data that support the findings of this study are openly available in the NCBI GenBank, accession numbers of all data are listed in this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen YH, Gan SS, Shao LL, Cheng CH, Hao JS. 2014a. The complete mitochondrial genome of the Pazala timur (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA Part A. 27(1):533–534.
- Chen YH, Huang DY, Wang YL, Zhu CD, Hao JS. 2014b. The complete mitochondrial genome of the endangered Apollo butterfly, Parnassius apollo (Lepidoptera: Papilionidae) and its comparison to other Papilionidae species. J Asia-Pac Entomol. 17(4):663–671.
- Hu SJ, Condamine FL, Monastyrskii AL, Cotton AM. 2019. A new species of the Graphium (Pazala) mandarinus group from Central Vietnam (Lepidoptera: Papilionidae). Zootaxa. 4554 (1):286–300.
- Hu SJ, Cotton AM, Condamine FL, Duan K, Wang RJ, Hsu YF, Zhang X, Cao J. 2018. Revision of Pazala Moore, 1888: the Graphium (Pazala) mandarinus (Oberthür, 1879) group, with treatments of known taxa and descriptions of new species and new subspecies (Lepidoptera: Papilionidae). Zootaxa. 4441(3):401–446.
- Hu SJ, Ning T, Fu DY, Haack RA, Zhang Z, Chen DD, Ma XY, Ye H. 2013. Dispersal of the Japanese pine sawyer, Monochamus alternatus (Coleoptera: Cerambycidae), in mainland China as inferred from molecular data and associations to indices of human activity. PLOS One. 8 (2):e57568.
- Kim MI, Baek JY, Kim MJ, Jeong HC, Kim KG, Bae CH, Han YS, Jin BR, Kim I. 2009. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol Cells. 28 (4):347–363.
- Kumar S, Stecher G, Tamura K. 2016. Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Park JS, Cho Y, Kim MJ, Nam SH, Kim I. 2012. Description of complete mitochondrial genome of the black-veined white, Aporia crataegi (Lepidoptera: Papilionoidea), and comparison to papilionoid species. Journal of Asian-Pacific Entomology. 15 (3):331–341.
- Racheli T, Cotton AM. 2009. Guide to the butterflies of the Palearctic region. Papilionidae. Part I. Milano: Omnes Artes.
- Zhang HH, Duan K, Hu SJ. 2018. On the immature stages of Graphium (Pazala) confucius Hu, Duan & Cotton, 2018 observed in Kunming, Yunnan, China. Butterflies. 79:18–25.