Abstract
Polygala tenuifolia Willd. is an Oriental plant that is widely distributed in East Asia; but the medicinal plant is a critically endangered species in Korea, where only a few individuals exist. In spite of that, there have been no molecular conservation studies on P. tenuifolia. In this study, we elucidated the complete chloroplast (cp) genome of P. tenuifolia and investigated its phylogenetic position based on the cp genomes of related taxa. Results showed the cp genome was 165,423 bp in length, consisting of a large single-copy region of 83,699 bp, a small single-copy region of 8,044 bp, and two inverted repeat regions of 36,840 bp. In addition, the plastid genome contained 123 genes, including 85 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. The present study is the first to report the complete cp genome of genus Polygala, information which may be valuable for future molecular phylogenetic and conservation studies on P. tenuifolia.
Polygala tenuifolia Willd., (order Fabales, family Polygalaceae) is a well-known traditional medicinal plant in East Asia (Chen et al. Citation2008), where it is widely distributed throughout the sunny grasslands, shrub forests, and thickets on the mountain slopes of China, Korea, Russia, and Mongolia (Kim Citation2007; Chen et al. Citation2008). On the Korean Peninsula, this herbaceous plant is confined to a limited region on a mountain in Andong-si, Gyeungsangbuk-do, where only a few individuals exist. Thus, it is considered a ‘critically endangered’ species in the Rare Plant Data Book in Korea (Korea Forest Service Citation2008). Nevertheless, there are no current conservation efforts for P. tenuifolia; and except to only a few chemical studies (Park et al. Citation2002), there have been exceedingly few genetic studies on the species. Thus, we aim to provide useful genetic information on P. tenuifolia by elucidating the complete chloroplast (cp) genome of this endangered species.
For chloroplast genomic analysis, fresh young leaf material was collected from natural populations of P. tenuifolia in South Korea (36°31′N, 128°28′E); a voucher specimen (Lee 18100) was deposited in the herbarium at Baekdudaegan National Arboretum. Total genomic DNA was sequenced using the Illumina MiSeq platform (LAS, Seoul, Korea). Prior to genome assembly, all raw sequence reads of the species discovered by Illumina were trimmed by Trimmomatic 0.32 (Bolger et al. Citation2014). The trimmed raw reads were assembled by a de novo method using Geneious R7 (Drummond et al. Citation2011). The cp genome sequence was mapped with respect to that of Ceratonia siliqua (KJ468096), and annotated using Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) and tRNAscan-SE (Lowe and Chan Citation2016). Afterward, the sequence was deposited in the National Center for Biotechnology Information GenBank under the accession number MT221251.
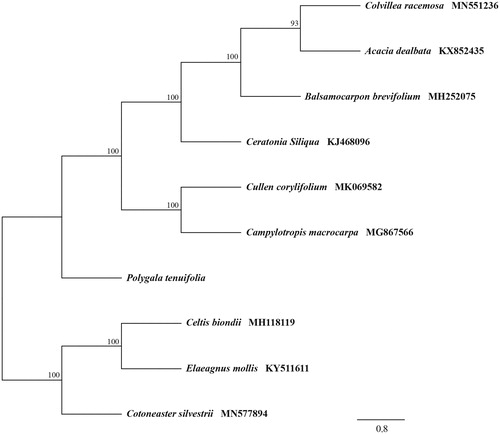
For phylogenetic analysis, we reconstructed the phylogeny using plastomes of the related nine taxa. The sequences were aligned by utilizing Multiple Alignment using Fast Fourier Transform (MAFFT) v.7.309 (Katoh and Standley Citation2013), and the tree was analyzed using Randomized Axelerated Maximum Likelihood (RAxML) 8.2.11 (Stamatakis Citation2014) with 1000 bootstrap replicates.
Results showed the plastid genome of P. tenuifolia was 165,423 bp in size, including a large-single copy (83,699 bp) region, a small-single copy (8,044 bp) region, and a pair of inverted repeat (36,840 bp each) regions. Furthermore, the circular genome contained 123 genes, including 85 protein-coding genes, 8 ribosomal RNA genes, and 38 transfer RNA genes. The overall guanine-cytosine content was 36.70%. Maximum-likelihood analysis showed P. tenuifolia clustered with other Fabales taxa under a strong bootstrap (). These data provide valuable genetic information that may assist in conservation efforts for the species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, J. G. Byeon, upon reasonable request
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, et al. 2011. GENEIOUS Pro v 7. http://www.geneious.com.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Kim YD. 2007. Polygalaceae. In: Flora of Korea Editorial Committee, editor. The Genera of vascular plants of Korea. Seoul: Academy Publishing Co. p. 829–832.
- Korea Forest Service. 2008. Rare plants data book in Korea. Pocheon: Korea National Arboretum; vol. 106. (in Korean).
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Park CH, Choi SH, Koo JW, Seo JH, Kim HS, Jeong SJ, Suh YH. 2002. Novel cognitive improving and neuroprotective activities of Polygala tenuifolia Willdenow extract, BT‐11. J Neurosci Res. 70(3):484–492.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Chen SK, Ma HY, Parnell J. 2008. Polygalaceae. In: WU ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 11. St. Louis (MO): Science Press, Beijing and Missouri Botanic Garden Press.