Abstract
Wolffia globosa is the smallest angiosperm in the world and can be found in Asia and parts of America. Also, it is commonly used as food in Southeast Asia. In this study, the complete chloroplast genome of the Wolffia globosa was assembled from the whole genome Illumina sequencing data. The assembled genome size is 169,405 bp in length, which composed of a large single copy region (LSC) of 92,171 bp, a small single copy (SSC) regions of 13,570 bp and separated by a pair of inverted repeat (IR) regions of 31,810 bp each. It encodes a total of 113 genes, including 78 protein-coding genes, 31 tRNA genes, and 4 rRNA genes. There are 22 duplicated genes in the predicted gene catalog. The overall GC content is 35.9% while the GC content of the LSC, SSC, and IR regions are 33.8%, 31.1%, and 40.0%, separately. Based on Bayesian phylogenetic analysis, it represents that Wolffia globosa was closely related to Wolffia australiana.
Main
Wolffia globosa is the smallest angiosperm in the world and belongs to the Araceae family as one of the 11 members of the Wolffia genus (Landolt Citation1994; Hoang et al. Citation2019). It is found in Asia such as China, Japan, Thailand, Indonesia, India, Malaysia, and Nepal, and parts of America such as the USA, Colombia, and Venezuela (Bog et al. Citation2013). Wolffia globosa is commonly found on the surface of calm water such as lakes, ponds, ditches, paddy fields and wetlands (Xu et al. Citation2015) during spring, summer and fall (Cheng and Stomp Citation2009). Because of this, it wasn’t readily regarded as an ingredient in human food in the past. However, now it is found that Wolffia globosa contains more protein with essential amino acids, fats and crude oil than beans, which is also used as a soybean substitute (Chantiratikul et al. Citation2010). Especially in Southeast Asia, Thailand, Wolffia globosa is called ‘khai nam’, ‘kai pum’ and ‘kai nahe’ are used as food ingredients such as salads, omelets and vegetable curries (Appenroth et al. Citation2018). Therefore Wolffia species provides an opportunity to reduce starvation caused by increasing worldwide food prices (UN Global Issues Citation2019). So Wolffia globosa is being tried to easily grow and eat at home with controlled environment agriculture (CEA) system (Ruekaewma et al. Citation2015). It is possible because Wolffia globosa has been reported to grown well on any water environment with 17.5–30 °C (Culley et al. Citation1981) and pH6.5–7.5 (Leng et al Citation1995).
The goal of the study was to facilitate and contribute to the genetic research of the same genus with Wolffia globosa. Therefore, for this study, the Whole genome Illumina sequencing data was assembled to use the complete chloroplast genome of Wolffia globosa. A phylogenetic analysis was also conducted, which can provide valuable information for identification and utilization of Araceae family.
Wolffia globosa was kindly provided by Rutgers Duckweed Stock Cooperative (RDSC; http://www.ruduckweed.org/), we used the Wolffia globosa accession of 8692, which was collected from the city of Chikugo in Fukuoka, Kyushu, Japan, located at 33°12′46.0″N, 130°29′54.6″E. Total genomic DNA was isolated from an individual of Wolffia globosa using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced by Illumina Hiseq Platform. The complete chloroplast genome was de novo assembled by the Novoplasty program of 2.7.2 version, using Ribulose bisphosphate carboxylase (RUBP) of the same genus Wolffia australiana as the seed (Dierckxsens et al. Citation2017). After that, it was annotated with Chlorobox’s GeSeq using the chloroplast references of Wolffia australiana, Lemna minor, Spirodela polyrhiza, Wolffiella linqulata of Lemnoideae that are the same subfamily with Wolffia globosa (Tillich et al. Citation2017). The complete chloroplast sequence with gene annotations was submitted to GenBank with the accession number MN881100.
The complete circular chloroplast genome of Wolffia globosa is 169,405 bp size, consisting of a large single copy region (LSC) of 92,171 bp, a small copy region (SSC) of 13,570 bp, and a pair of inverted repeat regions (IR) of 31,810 bp respectively. It encodes a total of 113 genes, including 78 protein-coding genes, 31 tRNA genes and 4 rRNA genes. Especially, there are a total of 22 double-duplicated genes, including 8 protein-coding genes, 8 tRNA genes, and 4 rRNA genes among those genes. The observations were very similar to Wolffia australiana, the same genus with Wolffia globosa. It is because all Wolffia globosa genes, except 2 protein-coding genes (rps12, pbf1) and 2 tRNA genes (trnS-CGA, trnG-GCC), are equally present in Wolffia australiana. The total GC content is 35.9% and the values of GC content in the LSC, SSC and IR regions are 33.8%, 31.1% and 40.0%, respectively.
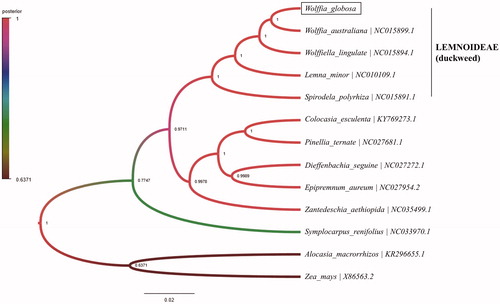
Phylogenetic analysis was also conducted to identify the phylogenetic location of Wolffia globosa. A phylogenetic tree was generated using the family Araceae by the software Beast v1.10.4 to conduct Bayesian-based evolutionary analysis (Suchard et al. Citation2018), which showed Wolffia australiana clustered into a clade with Wolffia globosa (). We expect that these findings would provide the basis for further investigation of chloroplast genome evolution in Wolffia.
Figure 1. Phylogeny of 12 species in Araceae and outgroup of zea mays based on Bayesian analysis of protein-coding sequences of chloroplasts. The cpREV + G + I model was used as the most suitable amino acid substitution model as a result of prottest v3.4.2. You can check the location of Wolffia globosa (GenBank accession number: MN881100).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number MN881100.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Appenroth K-J, Sree KS, Bog M, Ecker J, Seeliger C, Böhm V, Lorkowski S, Sommer K, Vetter W, Tolzin-Banasch K, et al. 2018. Nutritional value of the duckweed species of the genus Wolffia (Lemnaceae) as human food. Front Chem. 6:483.
- Bog M, Schneider P, Hellwig F, Sachse S, Kochieva EZ, Martyrosian E, Landolt E, Appenroth K-J. 2013. Genetic characterization and barcoding of taxa in the genus Wolffia Horkel ex Schleid.(Lemnaceae) as revealed by two plastidic markers and amplified fragment length polymorphism (AFLP). Planta. 237(1):1–13.
- Chantiratikul A, Chantirati P, Sangdee A, Maneechote U, Bunchasak C, Chinrasri O. 2010. Performance and carcass characteristics of Japanese quails fed diets containing Wolffia Meal [Wolffia globosa (L). Wimm.] as a protein replacement for soybean meal. Int J Poultry Sci. 9(6):562–566.
- Cheng JJ, Stomp AM. 2009. Growing duckweed to recover nutrients from wastewaters and for production of fuel ethanol and animal feed. Clean Soil Air Water. 37(1):17–26.
- Culley DD, Jr, Rejmánková E, Květ J, Frye J. 1981. Production, chemical quality and use of duckweeds (Lemnaceae) in aquaculture, waste management, and animal feeds. J World Mariculture Soc. 12(2):27–49.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hoang PT, Schubert V, Meister A, Fuchs J, Schubert I. 2019. Variation in genome size, cell and nucleus volume, chromosome number and rDNA loci among duckweeds. Sci Rep. 9(1):1–13.
- Landolt E. 1994. Taxonomy and ecology of the section Wolffia of the genus Wolffia (Lemnaceae). Ber Geobot Inst ETH Stiftung Rübel Zürich. 60:137–151.
- Leng R, Stambolie J, Bell R. 1995. Duckweed-a potential high-protein feed resource for domestic animals and fish. Livestock Res Rural Dev. 7:36.
- Ruekaewma N, Piyatiratitivorakul S, Powtongsook S. 2015. Culture system for Wolffia globosa L.(Lemnaceae) for hygiene human food. Songklanakarin J Sci Technol. 37:575–580.
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4(1):vey016.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- UN Global Issues. 2019. Food. https://www.un.org/en/sections/issues-depth/food/index.html
- Xu Y, Ma S, Huang M, Peng M, Bog M, Sree KS, Appenroth K-J, Zhang J. 2015. Species distribution, genetic diversity and barcoding in the duckweed family (Lemnaceae). Hydrobiologia. 743(1):75–87.
