Abstract
Most of the ungulates of Pakistan are either threatened or endangered species and their solitary and inaccessible life style makes them difficult to study. Therefore, estimating biodiversity, monitoring illegal trades and detecting commercial food frauds involving these species is a challenge for zoologists and conservation biologists. Here, we have attempted to exploit the discriminating power of mitochondrial COI gene to identify and to generate barcodes of the wild ungulate species of conservational importance found in Pakistan. 86 specimens of 19 wild ungulate species found in Pakistan were analyzed for their COI sequences. This is the first generated molecular data for many of these endemic and nearly endemic species. Intra and interspecific distances revealed distinct barcode gap for each species and a Neighborhood-joining tree able to discriminate all species into their respective clades. In conclusion, mtCOI is a powerful discriminatory tool for the taxonomic classification of ungulates especially for species that are inaccessible and require noninvasive sampling.
Introduction
Ungulates form the most diverse and thriving group of mammals in the world. Their numerous benefits make them a significant part of human society as most of the valuable economic domesticated animals are either artiodactyls or perrisodactyls (Vaughan et al. Citation2011). They are well adapted to exploit variety of habitats like mountains, deserts, plains and aquatic regions (Feldhamer Citation2007). Landscape of Pakistan provides ideal habitat for 19 species of cetartiodactyls belonging to five representative families; Suidae, Cervidae, Moschidae, Bovidae and Platanistidae including some of the endemic and nearly endemic sub-species like Punjab urial (Ovis vignei punjabiensis), flared horned markhor (Capra falconeri falconeri), Sindh ibex (Capra aegagrus blythi) and Indus blind dolphin (Platanista gangetica minor) (Molur Citation2003).
The advancement in species identification research using molecular techniques, particularly DNA barcoding has been a great milestone in determining ungulate diversity worldwide. It has been applied to studies regarding biodiversity, phylogeny, food adulteration cases, illegal trades of ungulates and their manufactured products (Kumar et al. Citation2017; Zhong et al. Citation2017; Khan et al. Citation2018; Shukla et al. Citation2019; Bhaskar et al. Citation2020). However, most of the ungulate diversity of Pakistan has not yet been studied through mitogenome analysis and has been restricted to the pioneer morphological studies conducted by Schaller (Citation1980), Mirza (Citation1998), and Roberts (Citation1997). In the present research, we have attempted to exploit the discriminating power of mitochondrial COI gene to generate reference barcodes for wild ungulates inhabiting Pakistan. This would be the first generated molecular data for many of these species. DNA barcodes of these species together with DNA sequences of other ungulate species retrieved from NCBI GenBank would also aid in resolving the long-time phylogenetic discrepancies of ungulates. The resulting data would also greatly aid in future studies involving taxonomic classification of these species.
Materials and methods
Taxon sampling and preservation
A total of 86 specimens of the 19 extant wild ungulate species were collected from different regions of Pakistan. Each species had at least three specimens except goitered gazelle (Gazella subgutturosa) and marcopolo sheep (Ovis ammon polii), both of which had one sample each due to rareness of the species and remote locations of habitat (). Specimen were morphologically identified with the help of wildlife expert (Dr. Fakhar-i-abbas and Jibran Haider) and morphological keys provided in literature (Roberts Citation1997; Groves and Grubb Citation2011).
Table 1. Sampling details of Pakistani ungulate species used in this study, including accession numbers, source, and conservation status.
Fecal samples were collected from zoos and breeding centers and immediately preserved in 70% ethanol, while Muscle tissues, skin and hair samples were obtained from cadavers from trophy hunters (). The acquired samples were preserved in 70% ethanol or kept at −20° Celsius. The specimens were also deposited in the specimen bank of Center for Bioresource Research (CBR), Islamabad and given appropriate voucher IDs ().
DNA extraction, COI amplification and sequencing
Total genomic DNA of the collected specimen was extracted through a modified organic DNA extraction method (Sambrook et al. Citation2001). COI gene was amplified using universal vertebrate primers, VF1d (5′-TTCTCAACCAACCACAARGA(Y)AT(Y)GG-3′) and VR1d (5′-TAGACTTCTGGGTGGCCRAARAA(Y)CA-3′) (Cai et al. Citation2011). All PCR amplifications included a negative control reaction which lacked template DNA and were conducted in a Veriti Thermal Cycler (Applied Biosystems).
The PCR products were analyzed using 1.5% agarose gel electrophoresis and quantified by Nanodrop Spectrophotometer (ND-2000). After confirmation of successful PCR by measuring absorbance ratio at 260/280 nm, amplicons were sequenced by ABI sequencer using facility of Center of Excellence in Molecular Biology (CEMB) Lahore.
Sequence analysis
The generated sequences were analyzed by a sequence alignment editing software BioEdit v7.0.5 (Hall Citation1999) with manual proofreading, sequences were analyzed to minimize any chances of polymorphic bases before any further analysis. All generated sequences were aligned by ClustalW using MEGAX (Kumar et al. Citation2018) to study parsimony informative sites. Sequences were further identified by using species identification tools available in BOLD (Barcode of Life Data system) and through BLAST GenBank databases to validate their identifications. Furthermore, Sequence analysis tools of BOLD was also utilized for estimating intra and interspecific divergences, nearest neighbor (NN) distances as well as barcode gap. A neighbor-joining phylogenetic tree was generated using Jukes-Cantor model (Jukes and Cantor Citation1969) by BOLD analysis system (Taxon ID Tree).
The barcode data generated from the sequences of Pakistani species were submitted to NCBI GenBank through the barcode submission tool and their accession numbers are given in . These sequences were also deposited online in a dataset named UNGPK in Barcode of Life Data System.
Results
Successful sequencing resulted in 731-bp sequences of COI gene which were then trimmed to 656-bp readable fragments for further analyses. The comparison of newly generated barcode sequences of Pakistani ungulates against already reported sequences of NCBI GenBank revealed that almost all the sequences were 98–100% identical with their respective species. The details of BLAST results are given in table S2. However, the sequence of musk deer showed 93% similarity to other musk deer species (KY792714.1) indicating its novelty in both the GenBank database. The sequence of gray goral (Naemorhedus goral) found in Pakistan is recognized as a sub-specie named Naemorhedus goral bedfordi and had 97% similarity to Naemorhedus goral (KT878720.1) reported in NCBI. Moreover, six of our sequences, Capra aegagrus blythi, Moschus cupreus, Naemorhedus goral bedfordi, Capra falconeri megaceros, Ovis vignei blanfordi and Ovis vignei punjabiensis were new to the database with no previous record available in databases.
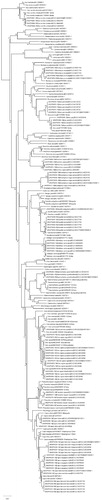
The intraspecific and interspecific divergences were calculated using MEGAX. Intraspecific divergence ranged from 0 within sheep (Ovis aries) and straight horned markhor (Capra falconeri megaceros) to 0.0246 within marcopolo sheep (Ovis ammon). The interspecific divergence ranged from 0.0025 between bearded pig (Sus barbatus) and javan warty pig (Sus verrrucosus) to 0.229 between Camelus dromedaries (Arabian camel) and Sus cebifrons (Visayan warty pig). According to our neighbor-joining tree (), the overall phylogeny of ungulates remained same as previous studies and it supported all the benchmark clades of ungulates forming similar split ups and clades (Hassanin et al. Citation2012; Wang and Yang, Citation2013).
Figure 1. Neighbor-Joining phylogenetic tree of ungulates using COI DNA barcodes. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 5). The analysis involved 188 nucleotide sequences. All positions containing gaps and missing data were eliminated.
Discussion
The present study was designed to develop DNA barcode data for the ungulates of Pakistan, most of which are either threatened or endangered. The data generated through this study will provide a reference to future studies related to ungulate species identification in Pakistan such as food fraudulent adulteration cases, illegal trade of wildlife and animal products, biodiversity and population studies. Several of the species are unique and have no previous records at Genbank or BOLD.
Barcoding provides researchers with vast opportunities to expand the taxonomic research for extant species that are not correctly identified by morphological techniques. Phenotypic similarities among closely related species or sub-species can be problem in assessing the identity of an organism on just morphological basis (Lorenz et al. Citation2005). Moreover, identification only through morphological means is often compromised in cases of commercial food frauds and illegal trade of wildlife and their products due to the extreme processing of these illegal products (Baker Citation2008).
The instances of utilizing DNA barcoding as a species identification tool are numerous. Bitanyi et al. (Citation2011) used a 470-bp fragment of COI gene to identify several species of Tanzanian antelopes. Yang et al. (Citation2015) rectified an issue of falsely identified musk deer population of Shanxi province in China using 672-bp region of COI gene. According to that study, a musk deer population identified as Moschus moschiferus in previous studies was actually M. berezovskii.
The overall sequence divergence patterns were consistent with many previous studies (Kumar et al. Citation2017; Bhaskar et al. Citation2020). Sequence divergence within each ungulate species was <1. Within family Moschidae, the lowest evolutionary distance was 0.01 between Anhui Musk Deer (Moschus anhuiensis) and Forest Musk Deer (Moschus berezovskii) while, highest sequence divergence 0.07 was recorded of Kashmir musk deer with Forest Musk Deer and Siberian Musk Deer (Moschus moschiferus).
Our study eliminated many misperceptions regarding the taxonomy of Pakistani ungulates. Built on the basis of morphological studies is a common perception that three sub-species of Markhor (Capra falconeri) inhabit in Pakistan, including Kashmir markhor (Capra falconeri cashmiriensis), flare horned markhor (Capra falconeri falconeri) and straight horned markhor (Capra f. megaceros) (Ali Citation2008; Ashraf et al. Citation2014). The percentage identity of Capra falconeri cashmiriensis was 99% to Capra falconeri falconeri and thus, it was inferred that both sub-species are in fact one which is also supported by morphological studies (Schaller and Khan Citation1975).
The classification of Urial (three sub-species in Pakistan) has also been controversial as some researchers report it as sub-species of Mouflon (Ovis orientalis) (Frisina et al. Citation2007; Ayaz et al. Citation2012), while some as Urial (Ovis vignei) (Awan et al. Citation2006; Siraj-ud-Din et al. Citation2018). Comparison of newly generated sequences of Urial with sequences already reported in GenBank and BOLD shows its high sequence similarity (99%) with Ovis vignei (KF938361.1) while only 97% similarity to Ovis orientalis (KF938360.1), therefore, we suggest that the sub-species of Urial found in Pakistan are sub-species of Ovis vignei rather than Asiatic mouflon (Ovis orientalis).
The Sindh ibex (Capra aegagrus blythi), locally known as Sara, was recognized as a sub-species of wild goat (Capra aegagrus aegagrus) by Hume (1875) on the basis of morphological characteristics. Comparative analysis of our Sindh ibex sequences against Genbank databases showed same similarity index with three different species of genus Capra viz. (98.34%) domestic goat (Capra hircus: AB736134.1), (98.33%) Markhor (Capra falconeri: MG742698.1) and (97.74%) bezoar (Capra aegagrus: KR059222.1). Neighbor-joining tree placed Sindh ibex as a sister clade to markhor (Capra falconeri falconeri) () although this relationship was not supported by high bootstrap value (49%). Similar results were observed in a previous study conducted by (Sultana et al. Citation2003) on the basis of mitochondrial Cytochrome b and D-loop genes.
Conclusion
We conclude that mitochondrial COI is an efficient marker for species identification as well as for studying the phylogenetic relationships of ungulates. However, our study also specifies the requirement of increased taxon sampling as well as studying more genes in order to resolve the taxonomic and phylogenetic issues that surround ungulates. In addition, because DNA barcoding contributes a lot in the conservation of endangered species and monitoring their illegal trades, we promote the utilization of DNA barcoding to devise suitable conservation strategies for the endangered ungulates of Pakistan.
Acknowledgements
This article is core part of doctoral thesis of A. Naseem. It is bilateral project between Center for Bioresource Research (CBR), Islamabad and Department of Zoology, University of Sargodha. We are thankful to Jibran Haider, District Forest Officer, Gilgit Baltistan for his help in provision of several specimens and their morphological identification.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ali S. 2008. Conservation and status of markhor (Capra falconeri) in the northern parts of North West Frontier Province, Pakistan [MS thesis]. Missoula: College of Forestry and Conservation, University of Montana.
- Ashraf N, Anwar M, Hussain I, Nawaz MA. 2014. Competition for food between the markhor and domestic goat in Chitral, Pakistan. Turk J Zool. 38(2):191–198.
- Awan GA, Festa-Bianchet M, Ahmad T. 2006. Poaching, recruitment and conservation of Punjab urial Ovis vignei punjabiensis. Wildlife Biol. 12(4):443–449.
- Ayaz S, Muhibullah AA, Jamil M, Khan MA, Qamar MF. 2012. Behaviour and biology of Ovis orientalis (urial) in Kotal Wildlife Park and Borraka Wildlife Sanctuary in Kohat. J. Anim. Pl. Sci. 22:29–31.
- Baker CS. 2008. A truer measure of the market: the molecular ecology of fisheries and wildlife trade. Mol Ecol. 17(18):3985–3998.
- Bhaskar R, Kanaparthi P, Sakthivel R. 2020. DNA barcode approaches to reveal interspecies genetic variation of Indian ungulates. Mitochondrial DNA B. 5(1):938–944.
- Bitanyi S, Bjørnstad GRO, Ernest EM, Nesje M, Kusiluka LJ, Keyyu JD, Mdegela RH, Røed KH. 2011. Species identification of Tanzanian antelopes using DNA barcoding. Mol Ecol Resour. 11(3):442–449.
- Cai Y, Zhang L, Shen F, Zhang W, Hou R, Yue B, Li J, Zhang Z. 2011. DNA barcoding of 18 species of Bovidae. Chin Sci Bull. 56(2):164–168.
- Feldhamer GA. 2007. Mammalogy: adaptation, diversity, ecology. Baltimore: JHU Press.
- Frisina MR, Awan GA, Woodford MH. 2007. Determining trophy harvest quotas through a status survey of urial (Ovis orientalis) in the Kalabagh Game Reserve, Punjab province, Pakistan. J Bombay Nat Hist Soci. 104(1):35.
- Groves C, Grubb P. 2011. Ungulate taxonomy. Baltimore: JHU Press.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic acids symposium series (Vol. 41, No. 41, pp. 95–98). London: Information Retrieval Ltd.; p. c1979–c2000.
- Hassanin A, Delsuc F, Ropiquet A, Hammer C, Jansen van Vuuren B, Matthee C, Ruiz-Garcia M, Catzeflis F, Areskoug V, Nguyen TT, et al. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. CR Biol. 335(1):32–50.
- Jukes TH, Cantor CR. 1969. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York (NY): Academic Press; p. 21–132.
- Khan FM, William K, Aruge S, Janjua S, Shah SA. 2018. Illegal product manufacturing and exportation from Pakistan: revealing the factuality of highly processed wildlife skin samples via DNA mini-barcoding. Nucleosides Nucleotides Nucleic Acids. 37(3):179–185.
- Kumar V, Sharma N, Sharma A. 2017. DNA barcoding of the Indian blackbuck (Antilope cervicapra) and their correlation with other closely related species. Egypt J Forensic Sci. 7(1):31.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lorenz JG, Jackson WE, Beck JC, Hanner R. 2005. The problems and promise of DNA barcodes for species diagnosis of primate biomaterials. Phil Trans R Soc B. 360(1462):1869–1877.
- Mirza ZB. 1998. Animal biodiversity of Pakistan. Pakistan: CERC.
- Molur S. 2003. Status and Red List of Pakistan’s mammals. Pakistan mammal conservation assessment & management plan workshop, 18–22 August 2003.
- Roberts TJ. 1997. The mammals of Pakistan. London: Ernest Benn.
- Sambrook J, Russell DW, Janssen K, Argentine J. 2001. Molecular cloning: a laboratory manual on the web. New York (NY): Cold Spring Harbor Laboratory.
- Schaller GB, Khan SA. 1975. Distribution and status of markhor (Capra falconeri). Biol Cons. 7(3):185–198.
- Schaller GB. 1980. Stones of silence. London: Andre Deutsch Limited.
- Shukla M, Joshi BD, Kumar VP, Thakur M, Mehta AK, Sathyakumar S, Goyal SP. 2019. Species dilemma of musk deer (Moschus spp) in India: molecular data on cytochrome c oxidase I suggests distinct genetic lineage in Uttarakhand compared to other Moschus species. Anim Biotechnol. 30(3):193–201.
- Siraj-Ud-Din M, Minhas RA, Ali U, Khan M, Awan MS, Shafi N, Ahmad B. 2018. Conservation status of Ladakh Urial (Ovis vignei vignei Blyth, 1841) in Gilgit Baltistan, Pakistan. PJZ. 50(1):1–400.
- Sultana S, Mannen H, Tsuji S. 2003. Mitochondrial DNA diversity of Pakistani goats. Animal Genetics. 34(6):417–421. doi:10.1046/j.0268-9146.2003.01040.x.
- Vaughan T, Ryan J, Czaplewski N. 2011. Mammalogy. London: Jones & Bartlett Learning.
- Wang Q, Yang C. 2013. The phylogeny of the Cetartiodactyla based on complete mitochondrial genomes. IJB. 5(3):30.
- Yang C, Xiao Z, Zou Y, Zhang X, Yang B, Hao Y, Moermond T, Yue B. 2015. DNA barcoding revises a misidentification on musk deer. Mitochondrial DNA. 26(4):605–612.
- Zhong W, Wang F, Li B, Jiang W, Yan H. 2017. Research on the non-directional test in meat adulteration based on DNA barcode. J Food Saf Qual. 8(5):1547–1551.
