Abstract
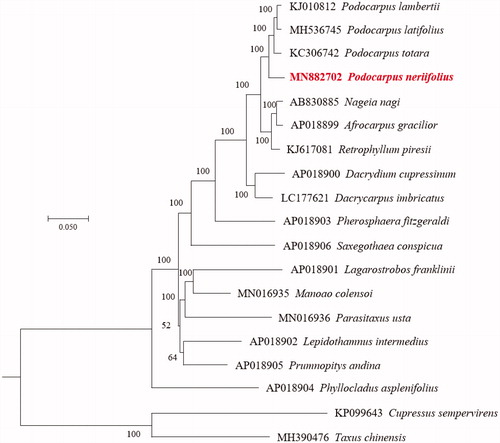
The complete chloroplast (cp) genome sequence of Podocarpus neriifolius D. Don was determined by BGISEQ-500 platform sequencing in this study. The entire cp genome was determined to be 133,878 bp in length. The genome contains a total of 119 encoding genes, including 82 protein-coding genes, four rRNA genes, and 33 tRNA genes. The overall GC content of P. neriifolius genome is 37.1%. A phylogenetic tree reconstructed by other 18 conifer species illustrated that P. neriifolius was the nearest relative to P. totara, P. latifolius, and P. lambertii.
Podocarpus neriifolius D. Don is a large evergreen tree mixing within evergreen broad-leaved forests. It is naturally distributed from east China to southwest China, but it was also recorded in southeast Asia and Nepal. P. neriifolius is a vital commercial tree for timber, medicine and garden (Shrestha et al. Citation2001; Van Sam et al. Citation2004; Wu et al. Citation2014). Moreover, this species has been listed as ‘Threatened Species’ in the IUCN Red List and as ‘Appendix III Species’ in The Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), which need urgent rescue action by the local government. Therefore, the aim of this study was to assemble the complete chloroplast (cp) genome of P. neriifolius according to BGISEQ-500 platform sequencing for phylogenetic studies and the conservation of genetic resources. The annotated cp genome of P. neriifolius has been uploaded to the GenBank with the accession number MN882702.
The sample of a single individual of P. neriifolius was collected from South China Botanical Garden, Chinese Academy of Sciences. The voucher specimen was stored at the Herbarium of Nanjing Forest Police College (accession no. 2019110102). The total genomic DNA (gDNA) was isolated from the young leaves of sample by the method of Doyle and Doyle (Citation1987), and whole genome sequencing on the BGISEQ-500 platform was conducted by Biodata Biotechnology Co. Ltd. (Hefei, China). In total, 33,686,206 clean reads were produced and then used for the de novo assembly with NOVOplasty 2.7.2 (Dierckxsens et al. Citation2016). Annotation was performed using the DOGMA (Wyman et al. Citation2004). There are 119 unique genes in the complete cp genome of P. neriifolius, including 33 tRNA, four rRNA genes, and 82 protein-coding genes. The overall GC content of the cp genome is 37.1% and this value in the protein-coding region, tRNA gene, rRNA gene, intron and intergenic region is 38.05, 42.78, 54.14, 37.04, and 32.21%, respectively.
To disclose taxonomic status of P. neriifolius, we reconstructed a maximum-likelihood (ML) tree using 21 single-copy homologous gene from other 18 gymnosperm taxa. The result of reconstructed phylogeny indicated that P. neriifolius was closer to P. totara, P. latifolius, and P. lambertii (). The genome information recorded here could be applied for the species conservation and the phyletic evolution of Podocarpaceae.
Acknowledgements
Data availability statement: The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MN882702.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JD. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Shrestha K, Banskota AH, Kodata S, Shrivastava SP, Strobel G, Gewali MB. 2001. An antiproliferative norditerpene dilactone, nagilactone C, from Podocarpus neriifolius. Phytomedicine. 8(6):489–491.
- Van Sam H, Nanthavong K, Kessler PJA. 2004. Trees of Laos and Vietnam: a field guide to 100 economically or ecologically important species. Blumea. 49(2):201–349.
- Wu J, Huang G, Zhan R, Liu Y, Chen YG. 2014. Study on the chemical constituents of Podocarpus neriifolius D. Don in Yunnan. J Hainan Normal Univ (Nat Sci). 27(1):36–38.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.