Abstract
Sanguisorba officinalis is well known for its incredible performance on stopping bleeding in herbal remedies in China. The chloroplast genome sequence of Sanguisorba officinalis was 155,479 bp length as the circular structure and typical quadripartite structure. It contained a large single-copy (LSC) region of 85,548 bp, a small single-copy (SSC) region of 18,769 bp and separated by two inverted repeat (IR) regions of 25,581 bp. The overall G + C content of the chloroplast genome sequence is 37.2%. This chloroplast genome contained 131genes that included 86 protein-coding (PC) genes, 37 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. In IR region, 18 genes was found, contained 7 PCGs species, 7 tRNA genes species and 4 rRNA genes species. In phylogenetic analysis, the result shown that Sanguisorba officinalis was closed relationship with Sanguisorba tenuifolia in the phylogenetic relationship using the Maximum-Likelihood (ML) method.
Sanguisorba officinalis is well known for its incredible performance on stopping bleeding in herbal remedies in China. It is distributed in the north temperate zone of Asia, mainly distributed in Asia, Europe, and North America countries, which grows in area at altitude of 30 meters to 3,000 meters (Meng et al. Citation2018). S. officinalis as one of the Chinese herb medicine is mainly composed active chemical components including triterpenoids, triterpenoid glycosides, lignans, lignans glycosides, polysaccharides and hydrolyze able tannins (Yang et al. Citation2016). It can protect the hematopoietic system, improve bone marrow micro-circulation, and adjust and improve body immunity and other functions in China (Eungyeong et al. Citation2018). But, we reported the chloroplast genome of Sanguisorba officinalis, which can accumulate data and information for further studies this species in phylogenetic relationship, and also can be applied to the further study the medicinal plants of S. officinalis in China.
The chloroplast DNA was extracted from the fresh samples of Sanguisorba officinalis that collected from the herb market near Beijing University of Chinese Medicin eat Beijing, China (116.47E, 39.97 N). The chloroplast DNA was the Plant Tissues Genomic DNA Extraction Kit (TIANGEN, BJ and CN) method and stored at School of Chinese Materia Medica, Beijing University of Chinese Medicine (No. BJUCM-01) that was purified and sequenced. We used the Fast QC software (Andrews Citation2015) to remove the low-quality reads for the quality control. And we used the MitoZ software (Meng et al. Citation2019) to assemble and annotate the chloroplast genome. The tRNA genes were further identified using tRNAs can-SE 2.0 (Lowe and Chan Citation2016). The physical map of the chloroplast genome of S. officinalis, we had been drawn using the OGDRAW software (Greiner et al.Citation2019). The chloroplast genome sequence had been submitted into GenBank (NCBI Accession No. MN045889).
The chloroplast genome sequence of Sanguisorba officinaliswas 155,479bplength as the circular structure that also has the typical quadripartite structure. It containeda large single-copy (LSC) region of 85,548 bp, a small single-copy (SSC) region of 18,769 bp and separated by two inverted repeat (IR) regions of 25,581 bp. The overall G + C content of the chloroplast genome sequence of S. officinalis is 37.2%, and while A of 31.0% (48,264 bp), T of 31.8% (49,387 bp), C of 18.9% (29,418 bp), G of 18.3% (28,410 bp). The chloroplast genome of S. officinalis contained 131genes that included86 protein-coding (PC) genes, 37 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. In IR region, 18 genes was found, contained7 PCGs species (rpl2, rpl23, ycf2, ndhB, rps7, rps12 and ycf1), 7 tRNA genes species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRN Agenes species (rrn16, rrn23, rrn4.5 and rrn5).
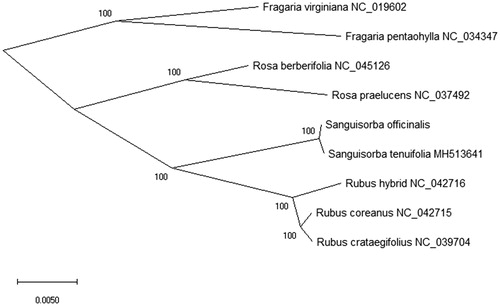
For the phylogenetic analysis, we assembled the plant species complete cp DNA sequences from NCBI and constructed the phylogenetic tree by the Maximum-Likelihood (ML) method using the GTR + G + I base substitution model. We used the MEGA X software (Kumar et al. Citation2018) to analysis the phylogenetic tree and performed using 5,000 bootstrap values replicate (). In phylogenetic analysis, the result shown that Sanguisorba officinalis was closed relationship with Sanguisorba tenuifolia (MH513641) in the phylogenetic relationship using the Maximum-Likelihood (ML) method. This research can be applied to the further study the medicinal plants of S. officinalis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data; [accessed 2020 Apr 15]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Eungyeong J, Soyoung K, Na-Rae L, Hayeon K, Sooin C, Chang-Woo H, Kim Y, Lee K, Kim B, Inn K, et al. 2018. Sanguisorba officinalis, extract, ziyuglycoside I, and II exhibit antiviral effects against hepatitis B virus. Euro J Integra Med. 20:165–172.
- Greiner S, Lehwark P, Bock R. 2019. Organellar Genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research. 47(W1):W59–64.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGAX: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evolut. 35(6):1547–1549.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Meng XX, Xian YF, Xiang L, Zhang D, Shi YH, Wu ML, Dong GQ, Ip SP, Lin ZX, Wu L, et al. 2018. Complete chloroplast genomes from Sanguisorba: identity and variation among four species. Molecules. 23(9):2137.
- Yang B, Hu J, Zhang F, Li J, Liu Q, Pu G, Liu H, Zhang Y. 2016. Herbal textural study on Sanguisorba officinalis L. Shandong University Tradit Chin Med. 5:412–414.