Abstract
The Chinese porcupine Hystrix hodgsoni belongs to the family Hystricidae, and is distributed in Asia. In this study, the total mitochondrial genome of H. hodgsoni was sequenced. The whole mitogenome is a typical circular DNA molecule of 16,833 bp and contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region, with a base composition of A 32.7%, G 13.9%, T 28.4%, and C 25.0%. Phylogenetic analysis indicated that Trichys fasciculata was the nearest sister to H. hodgsoni. The molecular data provided here would be useful for genetics study of H. hodgsoni.
The Chinese porcupine Hystrix hodgsoni is a species of rodent in the family Hystricidae and is found in Asia (Liu and Wu Citation2018). It occurs in a wide range of habitats, including primary and secondary forest, cultivated areas and plantations.
Limited mitochondrial genome sequences have been sequenced from the family Hystricidae, which comprises over 20 described extant species worldwide (Liu and Wu Citation2018). Artificial breeding of H. hodgsoni has been very popular in China. Here, we determined the complete mitochondrial genome of H. hodgsoni, with the adult individual collected from Tianyuan cultivation base (latitude: 27.509°N, longitude: 113.986°E), Pingxiang, Jiangxi Province. The specimen was deposited in Jiangxi Academy of Forestry with an accession number JXF-HZ-K293.
Total genome was extracted from muscle tissue using the standard phenol-chloroform protocol (Sambrook and Russell Citation2001). The mitochondrial genome sequence was obtained by Sanger sequencing.
The complete mitochondrial genome of H. hodgsoni (GenBank Accession Number MT240946; 16, 833 bp) contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region (Table S1). The whole base composition of the mitochondrial genome is showed as follows: A 32.7%, G 13.9%, T 28.4%, and C 25.0%; with an A + T-rich pattern of the vertebrate mitochondrial genomes (Tu et al. Citation2012; Huang and Tu Citation2016; Huang et al. Citation2016).
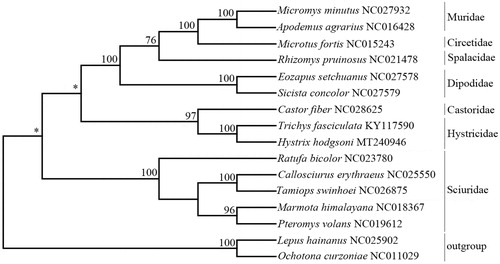
A phylogenetic tree () of mitochondrial genomes analyses of 13 species of Rodentia and two outgroup species, Ochotona curzoniae (NC_011029) and Lepus hainanus (NC_025902), was constructed based on the NJ method with the K2P model. Phylogenies (((Muridae, Circetidae) Spalacidae) Dipodidae) were recovered (), which was in concordant with the Yue et al. (Citation2015) study. Species of Sciuridae were clustered alone. Trichys fasciculata was the nearest sister to H. hodgsoni.
Figure 1. A phylogenetic tree (Neighbor-joining) based on 13 complete mitogenomes of species within Rodentia. Support values for each node were calculated from bootstrap values (BS). *Represents BS below 75.
So far, the species status of H. hodgsoni within Hystricidae is unclear. The complete mitochondrial genome provided here would be useful in systematics and genetics study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within its supplementary materials.
Additional information
Funding
References
- Huang ZH, Tu FY. 2016. Mitogenome of Fejervarya multistriata: a novel gene arrangement and its evolutionary implications. Genet Mol Res. 15(3):1–9.
- Huang ZH, Tu FY, Murphy RW. 2016. Analysis of the complete mitogenome of Oriental turtle dove (Strepopelia orientalis) and implications for species divergence. Biochem Syst Ecol. 65:209–213.
- Liu SY, Wu Y. 2018. Handbook of the mammals of China. Fuzhou: The Straits Publishing & Distributing Group.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd edn. New York: Cold Spring Harbour Laboratory Press.
- Tu FY, Fan ZX, Chen SD, Yin YH, Li P, Zhang XY, Liu SY, Yue BS. 2012. The complete mitochondrial genome sequence of the Gracile shrew mole, Uropsilus gracilis (Soricomorpha: Talpidae). Mitochondrial DNA. 23(5):382–384.
- Yue H, Yan CC, Tu FY, Yang CZ, Ma WQ, Fan ZX, Song ZB, Owens J, Liu SY, Zhang XY. 2015. Two novel mitogenomes of Dipodidae species and phylogeny of Rodentia inferred from the complete mitogenomes. Biochem Syst Ecol. 60:123–130.
