Abstract
Bupleuri Radix (Chaihu) is derived from two herbal materials, Bupleurum chinense DC. and B. scorzonerifolium Willd. In this study, we constructed and annotated a complete circular chloroplast (cp) genome of B. scorzonerifolium. The cp genome of B. scorzonerifolium is 155,824 bp in length, including two inverted repeat (IR) regions of 26,317 bp, and the large single-copy (LSC) region is 85,591bp, while the small single-copy (SSC) region is 17,601 bp. The GC content of whole cp genome is 37.7%. 113 different genes were annotated in the genome, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis suggested that B. Scorzonerifolium and other species of Bupleurum formed a monophyletic clade, and the genus Bupleurum had deep development system in Apiaceae.
Bupleuri Radix (Chaihu) is derived from two herbal materials, Bupleurum. chinense and B. scorzonerifolium (National Commission of Chinese Pharmacopoeia 2015).(National Commission of Chinese Pharmacopoeia. Citation2015) As a kind of traditional Chinese material medica, many medicinal plants are a major feature of traditional Chinese medicine. The roots of B. chinense and B. scorzonerifolium are the authentic basis of Bupleuri Radix. Bupleurum chinense’s roots are called ‘black Chaihu’ because of its black color, while B. Scorzonerifolium’s are called ‘red Chaihu’. In fact, this method of resolving material medica by color is very subjective and is difficult to identify many excessive forms. However, the pharmacological effects of B. chinense and B. scorzonerifolium are not exactly the same. They have different safety and poisoning doses. Different dynasties or periods used different species of Bupleurum in China. During the Song Dynasty in China, it was said that 9/10 of the decoctions have Bupleuri Radix. In addition to relieving liver and relieving depression, Chaihu has the effect of nourishing liver and yin. At that time, the amount of B. scorzonerifolium was relatively large, but the safety of B. scorzonerifolium was low.
Bioactive saponin compounds were emphasized, but they could not be used to distinguish B. chinense and B. Scorzonerifolium (Huang et al. Citation2009; Tian et al. Citation2009). It can be seen that it is necessary to distinguish B. chinense and B. Scorzonerifolium with gene.
Fresh samples of B. scorzonerifolium were collected from Zhangbei couty, Hebei province, (114°21′39.7″E, 41°25′11.4″N, 1396.7 m). Voucher specimen of B. scorzonerifolium was preserved in the herbarium of the Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences (Yongwanglu street 27, Beijing city). The specimen voucher number is 130722LY0211. The total plant DNA was extracted from about 0.2 g of leaves of B. Scorzonerifolium using the modified cetyltrimethylammonium bromide (CTAB) method (Allen et al. Citation2006).
The primer Tric-LAcp01F, Tric-LAcp01R, etc., were used to perform long-range PCR amplification. The length of the amplified fragment is 5–10 kp. Approximately 130 μL of each PCR product mixture was disrupted into fragments of about 100 bp with 1 minute and 50 seconds of sonication. 50 μL of ultrasonic disrupted products were taken from each sample and 6 × bromophenol blue was added to each agarose gel electrophoresis (2% gel, 110 V, 50 min), cut the gel to recover the fragment with a length of 400–500 bp, and the agarose gel purification and recovery kit is a new rapid extraction reagent for plant genomic DNA produced by Beijing Jinbaite Biotechnology.
The pairwise identity is 98.7% familiar to B. chinense (Zhang et al. Citation2019). AAA codon (1052 times) and ATT codon (1033 times) are most common. For B. Chinense and other plant species, all of the ribosomal RNAs are in the IR regions, suggesting that the IRs play an eminent role in the existence of the plastid in B. scorzonerifolium. Each sample has about 2 million reads of high-throughput sequencing one-way raw data. SPAdes software and Python script are used for splicing (Nurk et al. Citation2013), and the k-mer parameter is set to 95. The GenBank accession number for B. scorzonerifolium chloropalst complete genome is MT239475.
A conserved quadripartite structure was annotated with the overall GC content of 37.7%. The cp genome of B. scorzonerifolium is 155,824 bp in length, including two inverted repeat (IR) regions of 26,317 bp, and the large single-copy (LSC) region is 85,591bp, while the small single-copy (SSC) region is 17,601 bp. 113 different genes were annotated in the genome, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
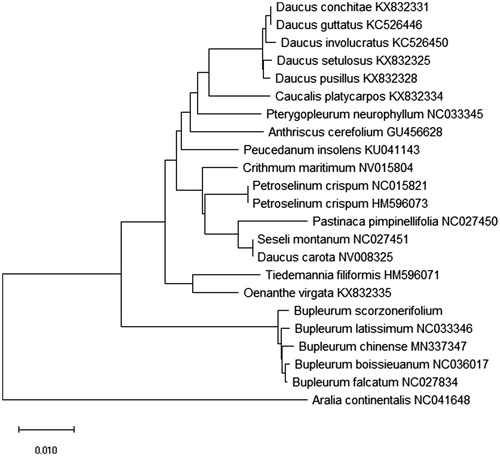
Arilia contientails (Araliaceae) was as the out group, and 21 species of Apiaceae were chosen to establish a neighbor-joining phylogenetic tree (Kumar et al. Citation2018, ) . The phylogenetic tree showed that the affinity of B. chinense and B. scorzonerifolium might not be near. The complete cp genome of B. scorzonerifolium provided great quantity genetic information for Araliaceae species conservation and identification. For safe use of the medical plants, the complete cp genome of B. scorzonerifolium would be significative with gene evidence.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethyl-ammonium bromide. Nat Protoc. 1(5):2320–2325.
- Huang HQ, Zhang X, Xu ZX, Su J, Yan SK, Zhang WD. 2009. Fast determination of saikosaponins in Bupleurum by rapid resolution liquid chromatography with evaporative light scattering detection. J Pharm Biomed Anal. 49(4):1048–1055.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- National Commission of Chinese Pharmacopoeia. 2015. Pharmacopoeia of the People’s Republic of China Vol. I. Beijing: Chemical Industry Press. (in Chinese).
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 20(10):714–737.
- Tian RT, Xie PS, Liu HP. 2009. Evaluation of traditional Chinese herbal medicine: chaihu (Bupleuri Radix) by both high-performance liquid chromatographic and high-performance thin-layer chromatographic fingerprint and chemometric analysis. J Chromatogr A. 1216(11):2150–2155.
- Zhang F, Zhao ZY, Yuan QJ, Chen SQ, Huang LQ. 2019. The complete chloroplast genome sequence of Bupleurum chinense DC. (Apiaceae), Mitochondrial DNA Part B. 4(2):3665–3666.