Abstract
Epimedium pubescens Maxim. is a well-known traditional Chinese medicine herb. In this study, the complete chloroplast genome of E. pubescens was sequenced. The genome was 158 956 bp in length, with a large single-copy region of 86,345 bp, a small single-copy region of 17,075 bp, and 2 inverted repeat regions of 27,768 bp. The genome consisted of 113 genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The GC contents were 38.82%. Phylogenetic analysis showed that E. pubescens of series Brachyrcerae was firstly clustered with E. acumiantum Franch. of ser. Dolichocerae, but not with E. brevicornu Maxim. from the same series.
Epimedium L. contains about 62 species and is the largest herbaceous genus of Berberidaceae (Ying et al. Citation2011; Y. Zhang et al. Citation2016; Y.J. Zhang et al. Citation2020). The overwhelming majority of Epimedium species are endemic to China, although some are found in eastern, southern, and central Asia as well as in Europe (Ma et al. Citation2011). More than 15 Epimedium species have been used as Chinese medicine and shown curative effects for sexual dysfunction, osteoporosis, cardiovascular diseases, asthma, menstrual irregularity, chronic nephritis, cancer, and so on (Jiang et al. Citation2015; Indran et al. Citation2016; Tan et al. Citation2016). However, Epimedium is a complex taxon and still has many questions on its infra-genetic phylogeny and species identification. Chloroplast genome has been widely utilized for reconstructing phylogenetic relationships and development of DNA barcodes and molecular markers for the identification of plant species/strains (Jansen et al. Citation2007; Avise Citation2009; Jung et al. Citation2014). In the previous studies, 16 Epimedium species have been reported (Lee et al. Citation2016; Zhanget al. Citation2016; Sun et al. Citation2018; Guo et al. Citation2019; Zhang et al. Citation2020). In the present paper, the complete chloroplast genome of Epimedium pubescens Maxim., one of the four original plants of Herba Epimedii in Chinese Pharmacopeia (The state Pharmacopoeia Committee of China Citation2015), was sequenced and the phylogenetic relationship of Epimedium was analyzed.
The chloroplast DNA of Epimedium pubescens was extracted from its fresh leaves materials which were collected in Dujiangyan, Sichuan, China (N31°0′11.65″; E103°36′32.51″). Voucher Yanjun Zhang 555 (HIB) was deposited in the Herbaria of Wuhan Botanical Garden, Chinese Academy of Sciences (HIB). A chloroplast genomic library was constructed with PCR technology and sequenced with Illumina Hiseq 2000 (Kim et al. Citation2017). High-quality reads were obtained with raw reads and assembled using NGS QC (Cai et al. Citation2015). Genome was assembled using CLC Genomics Workbench 11.0 software (CLC Bio, Aarhus, Denmark) and annotated using DOGMA (http://phylocluster.biosci.utexas.edu/dogma/) combined with the online alignment tools Blastx and ORF Finder (http://www.ncbi.nlm.nih.gov/).
The chloroplast genome sequence of Epimedium pubescens was submitted to NCBI, and the accession number was MN747095. The genome sequence was158,956 bp in length and the structure was a typical quadripartite, consisting of a large single-copy region with 86,345 bp (LSC), a small single-copy region with 17,075 bp (SSC), and two inverted repeat regions with 27,768 bp (IRs). The GC contents in the chloroplast genome were 38.82%. The chloroplast genome of E. pubescens contained 113 genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
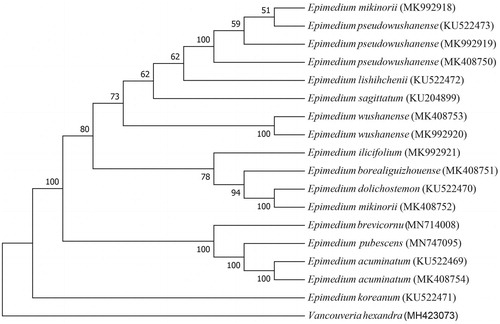
The maximum likelihood tree was constructed based on the genome sequences of Epimedium pubescens, 16 reported Epimedium species and Vancouveria hexandra as outgroup (Hansen et al. Citation2007) using MEGA7.0 (Kumar et al. Citation2016) (). The results were basically in accord with the previous phylogenetic trees based on chloroplast genome sequences of Epimedium (Y. Zhang et al. Citation2014, Citation2016; Y.J. Zhang et al. Citation2020). Epimedium pubescens of series Brachyrcerae was firstly clustered with E. acumiantum Franch. of ser. Dolichocerae, but not with E. brevicornu Maxim. from the same series. The phylogeny of Epimedium needs further study based on more chloroplast genome data of the genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study will be available in GenBank at https://www.ncbi.nlm.nih.gov/, Accession number MN747095.
Additional information
Funding
References
- Avise JC. 2009. Phylogeography: retrospect and prospect. J Biogeogr. 36(1):3–15.
- Cai J, Ma PF, Li HT, Li DZ. 2015. Complete plastid genome sequencing of four Tilia species (Malvaceae): a comparative analysis and phylogenetic implications. PLOS One. 10(11):e0142705.
- Guo ML, Ren L, Xu YQ, Liao BS, Song JY, Li Y, Mantri N, Guo BL, Chen SL, Pang XH. 2019. Development of plastid genomic resources for discrimination and classification of Epimedium wushanense (Berberidaceae). Int J Mol Sci. 20:4003.
- Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, Jansen RK. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 45(2):547–563.
- Indran IR, Liang RLZ, Min TE, Yong E-L. 2016. Preclinical studies and clinical evaluation of compounds from the genus Epimedium for osteoporosis and bone health. Pharmacol Therap. 162:188–205.
- Jansen RK, Cai ZQ, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Jiang J, Song J, Jia XB. 2015. Phytochemistry and ethnopharmacology of Epimedium L. Species Chin Herbal Med. 7(3):204–222.
- Jung J, Kim KH, Yang K, Bang KH, Yang TJ. 2014. Practical application of DNA markers for high-throughput authentication of Panaxginseng and Panax quinquefolius from commercial ginseng products. J Ginseng Res. 38(2):123–129.
- Kim SH, Cho CH, Yang M, Kim SC. 2017. The complete chloroplast genome sequence of the Japanese Camellia (Camellia japonica L.). Mitochondrial DNA Part B. 2(2):583–584.
- Kumar S, Stecher G, Tamura K. 2016. Mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lee JH, Kim K, Kim NR, Lee SC, Yang TJ, Kim YD. 2016. The complete chloroplast genome of a medicinal plant Epimedium koreanum Nakai (Berberidaceae). Mitochondrial DNA A. 27(6):4342–4343.
- Ma HP, He XR, Yang Y, Li MX, Hao DJ, Jia ZP. 2011. The genus Epimedium: an ethnopharmacological and phytochemical review. J Ethnopharmacol. 134 (3):519–541.
- Sun Y, Moore MJ, Landis JB, Lin N, Chen L, Deng T, Zhang J, Meng A, Zhang S, Tojibaev KS, et al. 2018. Plastome phylogenomics of the early-diverging eudicot family Berberidaceae. Mol Phylogenet Evol. 128:203–211.
- Tan HL, Chan KG, Pusparajah P, Saokaew S, Duangjai A, Lee LH, Goh BH. 2016. Anti-cancer properties of the naturally occurring aphrodisiacs: icariin and its derivatives. Front Pharmacol. 7:191.
- The State Pharmacopoeia Committee of China. 2015. The pharmacopoeia of the People’s Republic of China, Part 1. Vol. 167. Beijing, China: China Medical Science Press; p. 327–328.
- Ying TS, Boufford DE, Brach AR. 2011. Epimedium L. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 19. Beijing, China: Science Press; St. Louis, USA: Missouri Botanical Garden Press; p. 787–799.
- Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee S-C, Yang T-J, et al. 2016. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 7:306.
- Zhang YJ, Huang RQ, Wu L, Wang Y, Jin T, Liang Q. 2020. The complete chloroplast genome of Epimedium brevicornu Maxim (Berberidaceae), a traditional Chinese medicine herb. Mitochondrial DNA Part B. 5(1):588–590.
- Zhang Y, Yang L, Chen J, Sun W, Wang Y. 2014. Taxonomic and phylogenetic analysis of Epimedium L. based on amplified fragment length polymorphisms. Sci Hortic. 170:284–292.