Abstract
The first complete chloroplast genome sequence of Korean limestone endemic, Zabelia tyaihyoni, was reported in this study. The plastome size was 156,449 bp in total length, with one large single copy (LSC; 89,242 bp), one small single copy (SSC; 17,331 bp), and two inverted repeat (IR) regions (IRa and IRb, each with 24,938 bp). The overall GC content was 38.3% and the genome contained 128 genes, including 83 protein-coding with 3 pseudogenes (two ycf2 and accD), 37 transfer RNA and 8 ribosomal RNA genes. Phylogenetic analysis of 21 representative plastomes within the Caprifoliaceae suggests that Zabelia formed a monophyletic clade with 100% bootstrap value and Z. tyaihyoni was closely related to Z. biflora and Z. corymbosa.
Zabelia (Rehder) Makino (Makino Citation1948) is among one of the most well-known and important horticultural shrubs in the family Caprifoliaceae and is comprised of 5 to 10 species, occurring in warm temperate regions of eastern Asia (Hara Citation1983; Jacobs et al. Citation2010). In Korea, four taxa, Z. insularis, Z. biflora, Z. tyaihyoni, and Z. densipila, are currently recognized (Hisauchi and Hara Citation1954; Kim Citation2007; Hong et al. Citation2012). Z. tyaihyoni, named after the first Korean plant taxonomist, Professor Tyaihyon Chung (Ha Eun Herbarium, Sungkyunkwan University), is endemic to limestone area with very small population size in the central region (36°59′ ∼ 37°12′N) of the Korean peninsula (Kim et al. Citation2010; Chung et al. Citation2017). An extensive limestone mining has been resulted in significant habitat fragmentation and destruction, greatly threating population size and genetic resources. The newly characterized complete cp genome will provide indispensable genetic information to develop DNA markers for its population genetics and conservation biology in the future. We sequenced the complete plastome of Z. tyaihyoni and assessed its phylogenetic position within Caprifoliaceae.
Total DNA (Voucher specimen: 37°03′38″N 128°18′11″E, KNU-Lee180927005) was isolated using the DNeasy plant Mini Kit (Qiagen, Carlsbad, CA) and sequenced by the Illumina platform (Macrogen, Seoul, Korea). A total of 27,987,584 paired-end reads were obtained and assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008). The tRNAs were confirmed using tRNAsacn-SE (Lowe and Eddy Citation1997). The complete plastome length of Z. tyaihyoni (MT267474) was 156,449 bp, with one large single copy region (LSC; 89,242 bp), one small single copy region (SSC; 17,331 bp), and two inverted repeat regions (IRa and IRb; 24,938 bp each). The overall GC content was 38.3% and the plastome contained 128 genes, including 83 protein-coding genes with 3 pseudogenes (two ycf2 and accD), 8 rRNA, and 37 tRNA genes. A total of 14 genes were duplicated in the inverted repeat regions, including 7 tRNA, 4 rRNA, and 3 protein-coding genes.
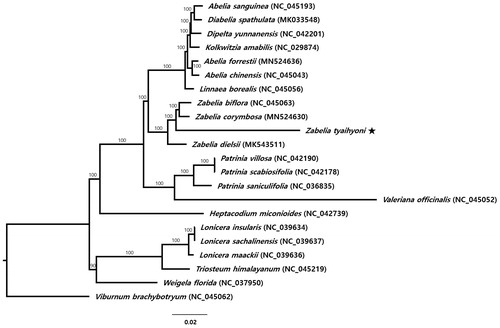
Twenty-one representative species of Caprifoliaceae, including Z. tyaihyoni, were aligned using MAFFT v.7 (Katoh and Standley Citation2013) and phylogenetic analysis was conducted using IQ-TREE v.1.6.7 (Nguyen et al. Citation2015). The genus Zabelia is a monophyletic group with 100% bootstrap support and is more closely related to the clade containing members of Linnaeoideae (Abelia, Diabelia, Dipelta, Kolkwitzia, and Linnaea) than to Valerianoideae (Patrinia and Valeriana) (). Within Zabelia, Z. tyaihyoni shows unusually long-branch length compared to its congeneric species and is closely related to Z. biflora and Z. corymbosa.
Data availability
The data that support the findings of this study are openly available in GenBank, National Center for for Biotechnology Information at (https://www.ncbi.nlm.nih.gov/genbank/), with reference number of MT267474.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chung GY, Chang KS, Chung JM, Choi HJ, Paik WK, Hyun JO. 2017. A checklist of endemic plants on the Korean peninsula. Korean J Pl Taxon. 47(3):264–288.
- Hara H. 1983. A revision of the Caprifoliaceae of Japan with reference to allied plants in other districts and the Adoxaceae. Tokyo: Academia Scientific Books.
- Hisauchi K, Hara H. 1954. On the genus Zabelia Makino. J Jpn Bot. 29:143–145.
- Hong MP, Kim YC, Nam GH, Lee BY. 2012. A new species of Zabelia (Linnaeaceae) from Korea. J Species Res. 1(1):1–3.
- Jacobs B, Pyck N, Smets E. 2010. Phylogeny of the Linnaea clade: are Abelia and Zabelia closely related? Mol Phyl Evol. 57(2):741–752.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim KA, Jang SK, Cheon KS, Seo WB, Yoo KO. 2010. Environmental and ecological characteristics of habitats of Abelia tyaihyoni Nakai. Korean J Pl Taxon. 40(3):135–144.
- Kim TJ. 2007. Zabelia. In: Flora of Korea Editorial Committee (ed.), The genera of vascular plants of Korea. Seoul: Academy Publishing Co. p. 935–936.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Makino T. 1948. On the genus Zabelia. Makinoa. 9:175.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.