Abstract
Rubus is an important genus, as a crucial Chinese traditional and Tibetan medicine, it has big ecological, economic, and social values; however, no plastid genome has been reported to date. Here we report species of this genus complete chloroplast genome of Rubus amabilis Focke. The chloroplast genome of R. amabilis Focke is found to be 155,279 bp in length with 37.31% GC contents. The cp genome sequences contained 131 genes, including 37 tRNA genes, 8 rRNA genes, and 86 mRNA genes, respectively. The complete chloroplast genome of these plants will be benefit for studies on the general characteristics and evolution of the Rubus family genome.
Rubus (Rubus L) is belonging to the diverse Rubus genus of Rosaceae, a large genus of about 750 kinds worldwide (Hancock Citation2008; Han and Liu Citation2009). According to the (Flora of China) recorded, there are probably 194 species in china (Editorial Committee of flora of China, Chinese Academy of Sciences Citation1985), and it is widely distributed in 27 provinces in China (Li et al. Citation2000). Rubus is a crucial Chinese traditional and Tibetan medicine, which has a long medicinal history, is also an important Mongolian medicine. In addition, the fruits of Rubus raspberries are a famous berry both at home and abroad, especially in abroad, the exploitation and utilization of Rubus are relatively mature. So, it is necessary to give close attention to these important species, a complete chloroplast genome data will contribute to the study and conservation of this plant. In this study, we assembled and characterized the complete chloroplast genome sequence of Rubus amabilis Focke using the genome skimming sequence data.
In this study, we reported the completed chloroplast genomes of R. amabilis Focke, the fresh leaves were collected from a plant in Qinghai Province, China (N34°29′4.30′′ and E102°08′19.10′′). The specimen was deposited at the herbarium of pharmacy department of Qinghai University, the specimen Accession number is 630224190520003LY. Genomic DNA was extracted following the modified CTBA method (Doyle Citation1987). The complete chloroplast genome was sequenced by Illumina HiSeq2500 platform (Illumina Inc., San Diego, CA), assembled with SPAdes version 3.10.1 (Bankevich et al. Citation2012) and annotated with CpGAVAS (Liu et al. Citation2012). Phylogenies trees were generated by maximum likelihood (ML) analysis. The sequences were multiple sequence alignment using MAFFT, choose the GTR and hill-climbing model, with the ML inference with 100 bootstrap replicates by RaxML-HPC2 on TG version 7.2.8 on the Cipres web server (Stamatakis et al. Citation2008). A General Time Reversible was selected as the best substitution models for the ML.
The complete chloroplast genome sequence of R. amabilis Focke (MN652918) was 155,279 bp in length. The typical quadripartite structure consists of a pair of IRs of 28,416 bp, an LSC region of 99,689 bp, and SSC region of 27,174 bp, respectively. The GC content of the chloroplast genomes was 37.31%. There were 131 predicted genes, 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
A total of 229 SSRs microsatellites were identified, and there were 148, 11, 59, 9 and 2 mono-, di-, tri-, tetra-, and penta-nucleotides repeats, respectively. Mononucleotide SSRs were the richest (64.63%). All the protein-coding genes presented a total of 26,696 codons, leucine (2813 codons, approximately 10.54% of the total) being the most abundant amino acid. Isoleucine (2257 codons, approximately 8.45%).
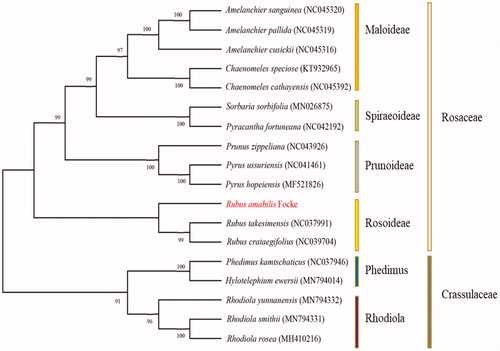
ML analysis showed that R. amabilis Focke formed a small branch with Rubus takesimensis and Rubus crataegifolius, belong to Rosoideae. It consisted in Rosaceae with Maloideae, Spiraeoideae, and Prunoideae (). The complete chloroplast date of R. amabilis Focke can provide reference for taxonomic study of this genus, and it also affords useful information for conservation and utilization of this multipurpose natural resources.
Data availability
The data that support the findings of this study are openly available in NCBI genbank at https://www.ncbi.nlm.nih.gov/nuccore/MN652918.1/, reference number MN652918.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Han J, Liu JW. 2009. The research progress on biological of Rubus. L. Chinese Wild Plant Resources. 28(2):1–49.
- Hancock JF. 2008. Temperate fruit crop breeding-chapter 12 raspberries. Berlin, Germany: Springer Science + Business Media B. V; p. 359–392.
- Li WL, He SA, Gu Y. 2000. An outline on utilization value of Chinese bramble (Rubus L.). J Wuh Bot Res. 18(3):237–243.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and genbank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Editorial Committee of flora of China, Chinese Academy of Sciences. 1985. Flora of China. Vol. 37. Beijing, China: Science Press.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.