Abstract
We determined the complete mitogenomes of two fiddler crabs Uca rosea (15,643 bp) and Uca paradussumieri (15,614 bp) of Ocypodidae within Brachyura. Both consist of 37 genes (13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes and one control region). The mitogenomes composition of U. rosea and U. paradussumieri are highly A + T biased, 70.77% and 71.12%, respectively, and showed positive AT (0.03 and 0.02) and negative GC (−0.29 and −0.24) skews. Phylogenetic analysis showing all the fiddler crabs were under monophyletic group relationship and belong to Ocypodidae; U. rosea has a sister group relationship with U. paradussumieri.
Fiddler crabs belong to the genus Uca (Decapoda, Brachyura, Ocypodidae), out of the 97 species of fiddler crabs identified, eight of them can be found in West Peninsular Malaysia with several species often found on sand or mudflat occupying different intertidal zones (Tan and Ng Citation1994; Rosenberg Citation2001). To date, more than 90 mitogenomes from species of the infraorder Brachyura have been sequenced; nevertheless, the evolution of Uca within Ocypodidae is still less described, and phylogeny has not been applied to study the evolution of fiddler crabs with respect to zonation and habitat. In this study, we analyzed the mitogenomes of Uca (Deltuca) rosea (Tweedie Citation1937) and Uca (Deltuca) paradussumieri (Bott Citation1973) and compared with other Ocypodidae and brachyuran.
The specimens (adult males U. rosea and U. paradussumieri) used in this study were collected at Tanjong Sepat, Selangor (2°35′N, 101°42′E) in October 2017. The collected specimen was transported and stored at Zoological Reference Museum Collection, Universiti Sains Malaysia (Voucher number:USMF3889 (U. rosea) and USMF3890 (U. paradussumieri)). Genomic DNA was extracted using Genomic-tip 100/G (Qiagen, Germany) and sequenced on a HiSeq 2500 platform (Illumina, USA). The clean data were then assembled using the NOVOPlasty software (Dierckxsens et al. Citation2017). The assembled complete mitogenomes are closed circular molecule of 15,643 bp (U. rosea, MN072632) and 15,718 bp (U. paradussumieri, MN072633) in length. Both consisted of 37 genes (13 PCGs, 22 tRNAs, two rRNAs and a control region), 23 out of 37 are encoded on the heavy (+) strand while 13 are located on the light (−) strand. The mitogenomes of U. rosea and paradussumieri are biased toward a higher representation of the nucleotides A and T (70.77 and 71.12%, respectively). All of the fiddler crabs mitogenomes show positive AT and negative GC skews, indicating a higher occurrence of A than T and C than G (Supplementary Table S2).
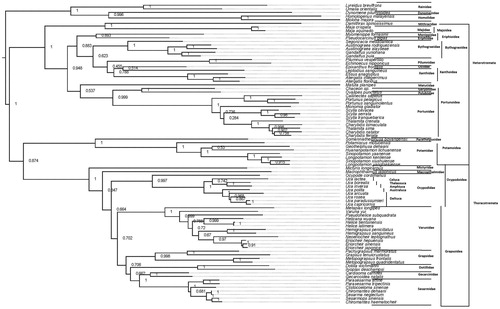
The complete mitogenome sequences and PCGs of other Brachyura (Supplementary Table S1) were downloaded from GenBank and used for phylogenetic analysis via the Maximum Likelihood (ML) method using MEGAX (Kumar et al. Citation2018). From the constructed phylogenetic tree using PCGs sequence (), U. rosea and U. paradussumieri belong to the Ocypodidae clade, the eight Ocypodidae fiddler crab constitute a strongly supported monophyletic clade (ML = 0.74 to 1.00) and branch from O. cardimanus with strong nodal value, ML = 0.997. The tree showed that the (U. polita + (U. arcuata + (U. rosea + (U. paradussumieri + U. capricornis)))) are in the same clade; while ((U. borealis + U. inversa) + U. lactea) are in sister clade. The phylogenetic tree (((((((((Sesarmidae + Gecarcinidae) + Dotillidae) + Grapsidae) + Varunidae) + Ocypodidae) + Macrophthalmidae) + Mictyridae) + (Potamidae) + Parathelpusidae)) was highly consistent with other studies, such as (((((Macrophtalmidae + Mictyridae) + Varunidae) + (Sesarmidae + Xenograpsidae) + Dotillidae) + Grapsidae) + Ocypodidae) (Guo et al. Citation2019) and ((((Xenograpsidae + Dotillidae) + Sesarmidae) + Gecarcinidae) + Ocypodidae (Wang et al. Citation2019).
Figure 1. Phylogenetic analysis of 86 Brachyura species based on concatenated amino acid sequence from 13 mitochondrial protein coding genes. The detailed list of species and accession number can be found in Supplementary Table S1. The tree was constructed using MEGAX (maximum-likelihood, JTT matrix based model, +G and complete deletion).
The data that support the findings of this study are openly available in [figshare.com] at https://doi.org/10.6084/m9.figshare.12082020
tmdn_a_1756942_sm3106.docx
Download MS Word (19.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bott R. 1973. Die Typus-Art der Gattung Uca Leach 1814 (Decapoda: Ocypodidae). Senckenbergiana Biologica. 54(4–6):311–314.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Guo H, Tang D, Wang Z, Shi X, Shen C, Cheng X, Ji C, Wang Z. 2019. Complete mitochondrial genome and phylogenetic analysis of Uca. Borealis Mitochondrial DNA B. 4(1):89–90.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Rosenberg M. 2001. The systematics and taxonomy of fiddler crabs: a phylogeny of the genus Uca. J Crustac Biol. 21(3):839–869.
- Tan CGS, Ng PKL. 1994. An annotated checklist of mangrove brachyuran crabs from Malaysia and Singapore. Hydrobiologia. 285(1–3):75–84.
- Tweedie MWF. 1937. On the crabs of the family Ocypodidae in the collection of the Raffles Museum. Bulletin of the Raffles Museum. 13:140–170.
- Wang Z, Shi X, Tao Y, Wu Q, Bai Y, Guo H, Tang D. 2019. Characterization of the complete mitochondrial genome of Uca lacteus and comparison with other Brachyuran crabs. Genomics. 111(4):799–807.
