Abstract
In this study, we report on the complete chloroplast genomes of Camellia perpetua and Camellia indochinensis for the first time. The complete chloroplast genome of C. perpetua is 156,804 bp in length and is comprised of two inverted repeat (IR) regions (25,996 bp each), a large single copy (LSC) region (86,570 bp), and a small single copy (SSC) region (18,242 bp). The chloroplast genome of C. perpetua harbors 86 protein-coding genes, 28 tRNA genes, and eight rRNA genes, with an overall GC content of 37.32%. The chloroplast genome of C. indochinensis showed similar characteristics as that of C. perpetua. The total length of C. indochinensis chloroplast genome is 156,571 bp, which includes 26,053 bp each of the two IR regions, 86,246 bp of the LSC region, and 18,219 bp of the SSC region. C. indochinensis chloroplast genome contains 85 protein-coding genes, 28 tRNA genes, and eight rRNA genes, with an overall GC content of 37.34%. Furthermore, a phylogenetic tree was constructed using the chloroplast genome sequences of the two species, and their phylogenetic positions were determined.
Camellia perpetua and Camellia indochinensis are evergreen shrubs belonging to the family Theaceae. C. perpetua is endemic to Chongzuo, Guangxi, China. It blooms throughout the year, producing bright yellow flowers, and serves as an excellent parental genotype in Camellia breeding programs (Huang et al. Citation2014). C. indochinensis also produces yellow colored flowers but shows a limited distribution in south Guangxi, China. C. indochinensis has been listed as a vulnerable (VU) species in the Threatened Species List of China’s Higher Plants (Qin et al. Citation2017).
Fresh leaves of C. perpetua and C. indochinensis were collected from the Nanning Golden Camellia Park (108.36°E, 22.82°N). Specimens of C. perpetua and C. indochinensis were placed in vouchers, labeled with unique codes (Pei Siyu 20191216 and Liu Shangli 20191216, respectively), and deposited at the herbarium of the Guangxi Institute of Botany (IBK). The fresh leaves of each species were placed in a centrifuge tube, rapidly frozen with liquid nitrogen, and submitted to the Shanghai Yuanxin Biological Company for DNA extraction, followed by paired-end sequencing using the Illumina HiSeq 400 platform. The filtered reads were assembled using NOVOPlasty (Dierckxsens et al. Citation2017) for sequence splicing, and annotated using Geneious (Kearse et al. Citation2012) and PGA (Qu et al. Citation2019).
The complete chloroplast genome of C. perpetua (GenBank accession MT157621) is 156,804 bp in length and includes two, inverted repeat (IR) regions (each 25,996 bp in length), a large single copy (LSC) region (86,570 bp), and a small single copy (SSC) region (18,242 bp), with an overall GC content of 37.32%. A total of 122 genes were annotated in C. perpetua chloroplast genome, including 86 protein-coding genes, 28 tRNA genes, and eight rRNA genes. The complete chloroplast genome of C. indochinensis (GenBank accession MT157620) showed similar characteristics, with a total length of 15,6571 bp, two IR regions of 26,053 bp each, an LSC region of 86,246 bp, and a SSC region of 18,219 bp. The chloroplast genome of C. indochinensis harbored 121 genes, including 85 protein-coding genes, 28 tRNA genes, and eight rRNA genes and showed an overall GC content of 37.34%. Notably, the ycf1 gene was absent in the chloroplast genome of C. indochinensis, unlike C. perpetua.
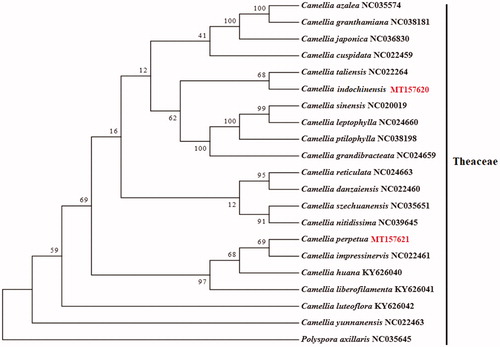
In this study, we sequenced the whole chloroplast genomes of C. perpetua and C. indochinensis for the first time using the Illumina Hiseq 400 platform. Additionally, to determine the phylogenetic position of C. perpetua and C. indochinensis in Theaceae. We constructed a maximum likelihood (ML) phylogenetic tree using chloroplast genome sequences of 21 Camellia species including C. perpetua and C. indochinensis. Chloroplast genomes of 18 representative Camellia species were compared with those of C. perpetua and C. indochinensis using MAFFT v.7 (Katoh and Standley Citation2013). A maximum-likelihood (ML) phylogenetic tree was constructed using RAxml program (Stamatakis Citation2014), with Polyspora axillaris as the outgroup. The results of phylogenetic analysis revealed that C. perpetua is most closely related to Camellia impressinervis, while C. indochinensis is most closely related to Camellia taliensis ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank, reference number [MT157620, MT157621]. These data were derived from the following resources available in the public domain: [constructed phylogenetic tree: NC035574, NC038181, NC036830, NC022459, NC022264, NC020019, NC024660, NC038198, NC024659, NC024663, NC022460, NC035651, NC039645, NC022461, KY626040, KY626041, KY626042, NC022463, NC035645, https://www.ncbi.nlm.nih.gov/genbank/]
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huang LD, Liang SY, Ye CX. 2014. A new species of Camellia from Guangxi, China. Guangdong Landscape Architect. 36:69–70.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Qin HN, Yang Y, Dong SY, He Q, Jia Y, Zhao LN, Yu SX, Liu HY, Liu B, Yan YH, et al. 2017. Threatened species list of China’s higher plants. Biodiversity Sci. 25(7):696–744.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.