Abstract
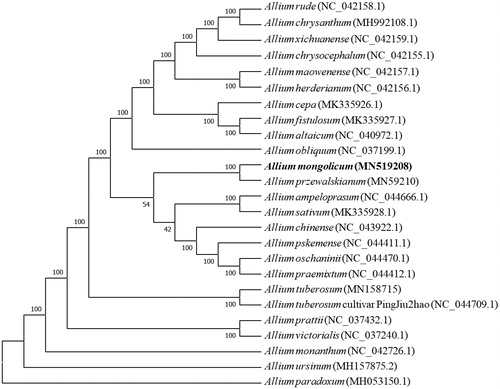
Allium mongolicum is a kind of wild vegetable with high nutritional value and even a traditional Chinese medicine. Here, we reported the complete chloroplast genome sequence of Allium mongolicum. The size of the chloroplast genome is 153,376 bp in length, including a large single copy region (LSC) of 82,912 bp, a small single copy region (SSC) of 18,054 bp, and a pair of inverted repeated regions of 26,205 bp. The Allium mongolicum chloroplast genome encodes 115 genes, including 69 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Phylogenetic tree showed that Allium mongolicum is closely related to Allium przewalskianum.
Allium mongolicum Regel, also known as the Mongolia leek, is a perennial and xerophytic herb. It grows in high altitude desert steppe and desert areas (Wang et al. Citation2013), mainly found in desert land in Qinghai, Gansu, Xinjiang and Inner Mongolia (Muqier et al. Citation2017). A. mongolicum has a higher nutritional value and medicinal value. Its ecological functions of sand-fixing, with good development prospects, cannot be underestimated (Wang et al. Citation2014). In this study, we assembled the complete chloroplast (cp) genome of A. mongolicum (Genbank accession number: MN519208) to provide genomic and genetic sources for further research.
The fresh leaves of A. mongolicum were collected from Gonghe (100.26E, 36.26 N), Qinghai Province, China. Total genomic DNA of A. mongolicum was extracted from leaf tissues with the modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen was deposited in Herbarium of the Northwest Institute of Plateau Biology (HNWP, whx2019003), Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Genome sequencing was achieved on the Illumina HiSeq Platform (Illumina, San Diego, CA) at Gene pioneer Biotechnologies Inc., Nanjing, China, and 6.92 GB of sequence data was generated. The trimmed reads were assembled via NOVOPlasty (Dierckxsens et al. Citation2017). The assembled genome was annotated using Plann v1.1 (Huang and Cronk Citation2015) and the annotation was corrected with Geneiousv11.0.3 (Kearse et al. Citation2012).
The chloroplast genome of A. mongolicum was 153,376 bp in length, containing a large single copy region (LSC) of 82,912 bp, a small single copy region (SSC) of 18,054 bp, and a pair of inverted repeat (IR) regions of 26,205 bp. Genome annotation predicted 115 genes, including 69 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The overall GC-content of the chloroplast genome was 36.06%, with the corresponding values in the LSC, SSC, and IR regions were 34.63%, 29.40% and 42.72%, respectively.
Phylogenetic analysis suggested that A. mongolicum is closely clustered with A. przewalskianum (), which was generated based on the 25 complete cp genomes. Alignment was conducted using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was built usingMEGA7 (Kumar Citation2016) with bootstrap set to 10,000. This study could lay a foundation for chloroplast genome engineering of A. mongolicum in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MN519208], or available from the corresponding author.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: De novo assembly of organelle genomes from whole genome DNA. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for anno-tating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. GeneiousBasic:an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.,
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Muqier Qi S, Wang T, Chen RW, Wang CF, Ao CJ. 2017. Effects of flavonoids from Allium mongolicum Regel on growth performance and growth-related hormones in meat sheep. Anim Nultr. 3:33–38.
- Wang GZ, Gao S, Wang XJ, Li J, Jia J. 2014. The research on physiological properties, functional ingredients, development and utilization of Allium mongolicum. Medicinal Plant. 5(1):67–69.
- Wang JK, Yang F, Zhao LH, Bao B. 2013. Analysis and comparison of nutritional components in Allium mongolicum Regel and Chinese chive. Acta Nutr Sin. 1:86–88.