Abstract
Quercus ciliaris C.C.Huang & Y.T.Chang is one of the dominant tree species in the subtropical evergreen broad-leaved forests of China. In this study, we sequenced and analyzed the complete chloroplast (cp) genome of the species. The circular genome is 160,842 bp in size, consisting of two copies of inverted repeat (IR) regions of 25,823 bp, one large single-copy (LSC) region of 90,294 bp, and one small single-copy (SSC) region of 18,902 bp. It encodes a total of 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis based on cp genome sequences of 35 Fagaceae species indicated that Q. ciliaris was among the members of section Cyclobalanopsis, and was most closely related to Q. myrsinifolia.
Quercus ciliaris C.C.Huang & Y.T.Chang is a medium to large-sized evergreen broadleaf tree in the beech family (Fagaceae). It is native to China where it grows at an altitude range of 500 to 2,600 m, and occurs widely across the Guizhou Plateau, the peripheral highlands of Sichuan Basin, and the mountainous areas of East China. Q. ciliaris is of great ecological importance for its dominant role in the subtropical evergreen broad-leaved forest. Furthermore, young leaves and acorns of the species are important foods for many animals, including threatened primates like golden snub-nosed monkey (Rhinopithecus roxellana) and Tibetan macaque (Macaca thibetana) (Li et al. Citation2013). Previous phylogenomic studies have revealed that Q. ciliaris belongs to the single-celled trichome base (STB) lineage within Quercus section Cyclobalanopsis (Deng et al. Citation2018). However, the plastid genome sequence of the species remains unknown. In this study, we report the complete chloroplast (cp) genome of Q. ciliaris based on Illumina paired-end sequencing data.
Fresh leaves were sampled from an adult tree near the Lansheng Bridge in the Huangshan Mountain Scenic Area, Anhui Province, China (30°6′0.95″N, 118°10′20.35″E, 642 m). The voucher specimen was deposited at the Herbarium of Nanjing Forestry University (accession number YL20190417007). Total DNA extraction and whole genome sequencing on the Illumina Hiseq X Ten platform were conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China). A total of 29,152,346 clean reads were produced and then used for the de novo assembly with NOVOplasty 2.7.2 (Dierckxsens et al. Citation2016). Gene annotation was performed using the CpGAVAS pipeline (Liu et al. Citation2012). The annotated cp genome was deposited in the GenBank database under the accession number MN199024.
The complete plastid genome of Q. ciliaris is a circular molecule of 160,842 bp in length, consisting of two copies of IR (25,823 bp) separated by the LSC (90,294 bp) and SSC (18,902 bp) regions. The cp genome encoded a total of 133 genes, of which 114 were unique and 19 were duplicated in the IR regions. The 114 unique genes contained 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Among those, nine protein-coding genes (rpoC1, ndhB, ndhA, rpl2, rpl16, petB, atpF, rps16, petD) and six tRNA genes (trnK-UUU, trnI-GAU, trnA-UGC, trnG-GCC, trnV-UAC, trnL-UAA) contained one intron, and three genes (rps12, ycf3, clpP) contained two introns. The overall GC content was 36.89%, while the corresponding values of the LSC, SSC, and IR regions were 34.74%, 31.09%, and 42.78%, respectively.
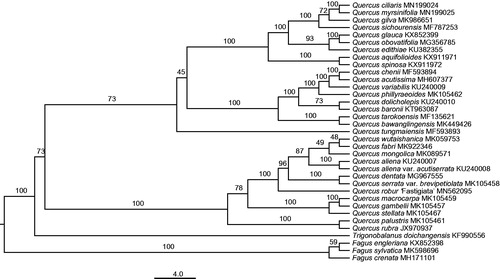
To identify the phylogenetic position of Q. ciliaris, a maximum-likelihood (ML) tree of 35 Fagaceae species was inferred under the GTRGAMMA substitution model using raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Species from Trigonobalanus and Fagus were selected as outgroups. Four locally collinear blocks were identified and a matrix of 92,127 bp was generated by the HomBlocks pipeline (Bi et al. Citation2018). Node support was assessed using 1,000 fast bootstrap replicates. Our results indicated that Q. ciliaris was among the members of section Cyclobalanopsis, and was most closely related to Q. myrsinifolia with 100% bootstrap support ().
Figure 1. The maximum-likelihood (ML) phylogenetic tree reconstructed by raxmlGUI 1.5 (Silvestro and Michalak Citation2012) based on cp genome sequences of 35 Fagaceae species. The bootstrap support value is labeled for each node.
Acknowledgments
The authors thank Jun Wu and Mingxia Tang from Garden Bureau, Mount Huangshan Scenic Area Management Committee for their help with fieldwork; Cheng Zhang from Nanjing Forestry University for his assistant with data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The complete chloroplast genome sequence of Quercus ciliaris is deposited in the GenBank database under the accession number MN199024.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Deng M, Jiang XL, Hipp AL, Manos PS, Hahn M. 2018. Phylogeny and biogeography of East Asian evergreen oaks (Quercus section Cyclobalanopsis; Fagaceae): insights into the Cenozoic history of evergreen broad-leaved forests in subtropical Asia. Mol Phylogenet Evol. 119:170–181.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:18.
- Li YK, Jiang ZG, Miao T. 2013. Diet and its seasonality of golden snub-nosed monkeys (Rhinopithecus roxellana) in Qingmuchuan Nature Reserve, Shaanxi Province, China. Acta Theriologica Sinica. 33:246–257. [Chinese]
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
