Abstract
Toxicodendron sylvestre (Siebold & Zucc.) Kuntze is a deciduous tree species in the poison ivy genus of the sumac family. In this study, we sequenced and analyzed the complete chloroplast (cp) genome of T. sylvestre. The circular genome is 159,600 bp in length, including two copies of inverted repeat (IR) regions of 26,465 bp, one large single-copy (LSC) region of 87,630 bp, and one small single-copy (SSC) region of 19,040 bp. It encodes a total of 113 unique genes, including 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on 17 cp genome sequences revealed that T. sylvestre was more closely related to T. succedaneum.
The poison ivy genus (Toxicodendron) is well-known for producing the skin-irritating oil urushiol that can cause severe allergic reactions to humans (Jiang et al. Citation2019). This genus has a disjunct distribution between eastern Asia and North America (Nie et al. Citation2009). Among its eastern Asian members, Toxicodendron sylvestre (Siebold & Zucc.) Kuntze is a deciduous tree species widely distributed across Japan, Korea, and south of the Yangtze River in China. It is also characterized by the dense hairs throughout the young branchlets, terminal buds, leaves, and inflorescences. Using five nuclear and chloroplast DNA regions, previous studies have shown that T. sylvestre was more closely related to T. succedaneum (Jiang et al. Citation2019). However, the plastid genome sequence of the species remains unknown. In this study, we assembled and annotated the complete chloroplast (cp) genome of T. sylvestre, which will provide beneficial genomic resources for the future studies on the evolutionary histories of Toxicodendron.
Fresh leaves of T. sylvestre were sampled from a wild individual growing in the Tianmu Mountain National Nature Reserve, Zhejiang Province, China (30.31°N, 119.44°E). The voucher specimen was stored at the Herbarium of Nanjing Forestry University (accession number LW190803001). The total DNA was extracted according to the previous report (Wang et al., Citation2020). The whole genome sequencing was conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China) on the Illumina Hiseq X Ten platform. A total of 21,984,629 clean reads were produced and used for the de novo assembly with NOVOplasty 2.7.2 (Dierckxsens et al. Citation2016). Gene annotation was performed using the CpGAVAS pipeline (Liu et al. Citation2012).
The circular complete cp genome of T. sylvestre (GenBank accession number MT211615) was 159,600 bp in length, and contained two copies of IR (26,465 bp), which were separated by one LSC (87,630 bp) region and one SSC (19,040 bp) region. The overall GC content was 37.85%, while the corresponding values of the LSC, SSC, and IR regions were 35.95%, 32.45%, and 42.94%, respectively. A total of 134 genes were encoded, of which 113 were unique and 21 were duplicated in the IR regions. The 113 unique genes contained 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Among those, three (rps12, ycf3, and clpP) contained two introns, and 16 (nine protein-coding genes and seven tRNA genes) contained one intron.
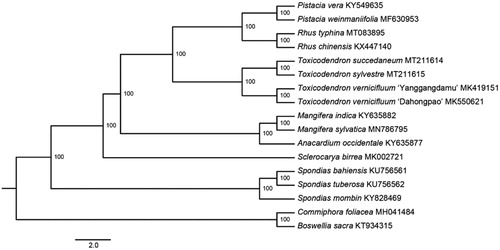
To identify the phylogenetic position of T. sylvestre, we reconstructed a maximum-likelihood (ML) tree using cp genome sequences of 15 Anacardiaceae species and two outgroups from Burseraceae (). Five locally collinear blocks (LCBs) were identified and a matrix of 90,009 bp was generated by the HomBlocks pipeline (Bi et al. Citation2018). The ML analyses were performed with RaxmlGUI 1.5b1 (Silvestro and Michalak Citation2012) under the GTRGAMMA model. Branch support values were estimated for each node based on 1000 samples of rapid bootstrap. Our results indicated that T. sylvestre was more closely related to T. succedaneum with 100% bootstrap support.
Acknowledgments
The authors thank Mingshui Zhao from the Tianmu Mountain National Nature Reserve of Zhejiang Province for his help with fieldwork; Cheng Zhang from Nanjing Forestry University for his assistant with data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence of Toxicodendron sylvestre is deposited in the GenBank database under the accession number MT211615.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:18.
- Jiang Y, Gao M, Meng Y, Wen J, Ge XJ, Nie ZL. 2019. The importance of the North Atlantic land bridges and eastern Asia in the post-Boreotropical biogeography of the Northern Hemisphere as revealed from the poison ivy genus (Toxicodendron, Anacardiaceae). Mol Phylogenet Evol. 139:106561.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Nie ZL, Sun H, Meng Y, Wen J. 2009. Phylogenetic analysis of Toxicodendron (Anacardiaceae) and its biogeographic implications on the evolution of north temperate and tropical intercontinental disjunctions. J Syst Evol. 47(5):416–430.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Wang L, He N, Li Y, Fang YM, Zhang F. 2020. Complete chloroplast genome sequence of Chinese lacquer tree (Toxicodendron vernicifluum, Anacardiaceae) and its phylogenetic significance. Biomed Res Int. 2020:9014873.