Abstract
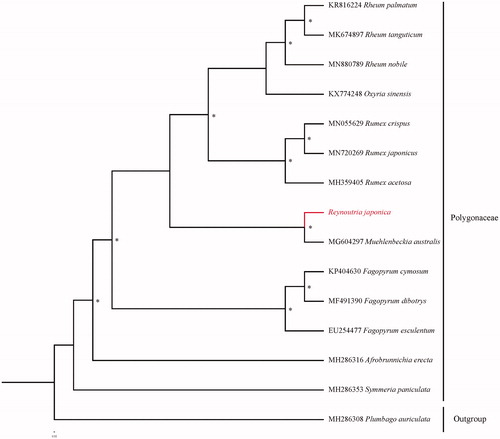
The complete chloroplast genome of medicinal Reynoutria japonica Houtt. was sequenced and assembled in this study. The genome is 163,410 bp in length and contained 129 encoded genes in total, including 86 protein-coding genes, 8 ribosomal RNA genes, and 36 transfer RNA genes. The phylogenetic tree based on 14 chloroplast genomes in Polygonaceae support that R. japonica is related to Muehlenbeckia.
Reynoutria japonica Houtt., native to East Asia, is a large perennial herb in Polygonaceae. It is commonly known as Asian knotweed or ‘Huzhang’ as a traditional Chinese medicine. In China and Japan, the root of R. japonica is used as effective agent used for the treatment of hyperlipemia, inflammation, infection and cancer, although there is no high-level evidence of any medical efficacy in clinical studies (Peng et al. Citation2013). More than 67 compounds were isolated and identified from this plant, such as piceid, resveratroloside, resveratrol, physcion, and emodin (Shan et al. Citation2008). Especially, the active compound emodin has recently been considered as a possible treatment for the COVID-19 (Yang et al. Citation2020). In this study, the first complete chloroplast genome of R. japonica was assembled and characterized and will provide organelle molecular basis for this medicinal important species.
The leaves of R. japonica were collected from Chun’an, Zhejiang, China (GPS: E118°46′57″, N29°50′45″, Voucher No. ZSTU02676, deposited at Zhejiang Sci-Tech University). DNA was extracted from its silica-dried leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA). The total genomic sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). In total, about 11.8 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. Then, NOVOPlasty (Dierckxsens et al. Citation2017) was used de novo to assemble the chloroplast genome with rbcL gene as seed. Following Liu et al (Citation2017, Citation2018), aligning and annotation were conducted by BLAST, GeSeq (Tillich et al. Citation2017) and GENEIOUS prime (Biomatters Ltd, Auckland, New Zealand).
The complete chloroplast genome of R. japonica (GenBank accession No. MT301955) has a length of 163,410 bp with a typical circle structure and a GC content of 37.5%. It consists of a large single-copy region (LSC, 875,95 bp, 35.6% GC content), a small single-copy region (SSC,135,67 bp, 32.8% GC content), and two inverted repeat regions (IR, 31124 bp, 41.2% GC content). In total, there were 129 genes in R. japonica, including 86 protein-coding genes, 8 rRNA genes, and 36 tRNA genes. Among them, seven protein-coding genes (rpl2, rpl23, ycf1, ycf2, ndhB, rps7, rps19), all four rRNA genes (rrn16, rrn23, rrn4.5, rrn5), and seven tRNA genes (trnH-CAU, trnL-CAA, trnV-GAC, trnL-CAU, trnA-UGC, trnR-ACG, trnN-GUU) have two copies. Five of these protein-coding genes (rps16, atpF, rpoC1, ndhB, ndhA) have one intron each and two (ycf3, clpP) of them have two introns.
Fourteen species with available chloroplast genomes were selected to study the phylogentic placement of R. japonica in Polygonaceae (). The sequence alignment was conducted with MAFFT v7.3 (Katoh and Standley Citation2013). The phylogenetic tree was drawn by the software IQtree (Nguyen et al. Citation2015) with 5000 bootstrap replicates and GTR + F+R3 model. The phylogenetic tree revealed that R. japonica was related to Muehlenbeckia in Polygonaceae, which was consistent with the previous study (Sanchez and Kron Citation2008).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The DNA matrix and phylogenetic tree that support the findings of this study are openly available in Github at https://github.com/andresqi/Reynoutria-japonica-chloroplast-genome
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Liu L, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu C. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of fagales. Front Plant Sci. 8:968.
- Liu L, Li P, Zhang HW, Worth J. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. Sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res-JPN. 23(5):318–323.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol E. 32(1):268–274.
- Peng W, Qin RX, Li XL, Zhou H. 2013. Botany, phytochemistry, pharmacology, and potential application of Polygonum cuspidatum Sieb.et Zucc.: a review. J Ethnopharmacol. 148(3):729–745.
- Sanchez A, Kron KA. 2008. Phylogenetics of polygonaceae with an emphasis on the evolution of Eriogonoideae. Syst Bot. 33(1):87–96.
- Shan B, Cai YZ, Brooks JD, Corke H. 2008. Antibacterial properties of Polygonum cuspidatum roots and their major bioactive constituents. Food Chem. 109(3):530–537.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yang L, Liu JQ, Bian Y, Chen M, Shuai XC, Tong RS, Tang XJ, Cai LL. 2020. The possibility of using emodin from Chinese herbal medicine as treatment for novel coronavirus pneumonia. Pharm Today. Advance online 1-19. (In Chinese).