Abstract
The Crinoidea is the ancestor class of echinoderm classes, whereas its evolution history remains the least studied in extant echinoderms. In this study, a complete 15,772 bp genome for the crinoid echinoderm, Florometra species (Echinodermata, Crinoidea) mitochondrion was assembled through Illumina HiSeq platform. The complete mitochondrial genome of Florometra sp. contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 3 regions of unassigned sequence (UAS) including one putative control region. Overall genomic structure and gene orientation are identical within the genus Florometra. Phylogenetic analysis using the maximum likelihood method validated the sister relationship with Florometra serratissima.
Keywords:
Extant crinoids such as feather stars and sea lilies constitute crucial components of all other living echinoderms, including Asteroidea, Crinoidea, Echinoidea, Holothruoidea, and Ophiuroidea (Foote Citation1999; Rouse et al., Citation2013). Crinoids are useful model animals for the understanding evolutionary history of echinoderms and other animal taxa, as they are long-lived history with a huge fossil record spanning nearly half a billion years (Hess et al. Citation1999; Wright et al. Citation2017). Although crinoids are one of the largest clades of echinoderms with complex diversity, research on their phylogenetic and evolutionary history remains to be explored due to a wide range of geographical distribution and insufficient genomic information. To date, two complete mitochondrial genomes (Florometra serratissima and Florometra sp.) were reported in the genus Florometra (Scouras and Smith Citation2001; Nam et al. Citation2020).
In this study, we sequenced the complete mitogenome of Florometra sp. (Accession no. MT302206). An individual of Florometra sp. was isolated at the aquaculture enclosure (Tongyeong, Gyungnam, South Korea; 34°51′N, 128°21′E). The voucher specimen was deposited in the Research Institute of Basic Sciences of Incheon National University (Specimen ID: 201805-Crinoid008). Genomic libraries were constructed from the total genomic DNA using the TruSeq RNA Sample Preparation Kit according to the manufacturer’s instructions (Illumina, San Diego, CA, USA) and sequenced [No. aligned reads: 15,984; coverage (X): 143.06] at a commercial company (Phyzen, Seoul, Republic of Korea). Additional PCR procedure was employed to confirm the DNA sequence of COI, cytB, and UAS I region. The mitogenome was annotated using the MITOS web-based software (Bernt et al. Citation2013) and detailed annotation was performed using NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
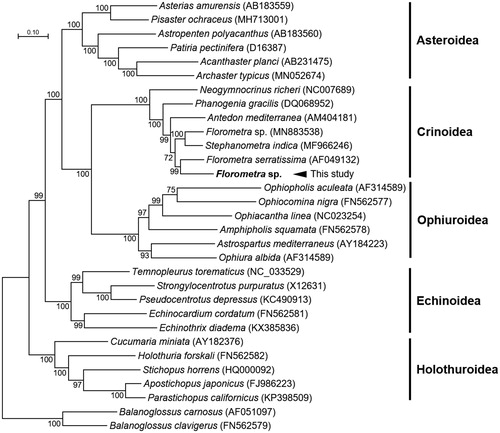
The complete mitogenome of Florometra sp. was 15,772 bp in length and contained a typical set of 13 PCGs, 22 tRNAs, 2 rRNAs, and 3 UASs, including 1 putative control region. Overall gene contents, their orientations, and the positions of tRNAs and three UASs were identical to the mitogenomes of the genus Florometra (Scouras and Smith Citation2001; Nam et al. Citation2020). The crinoid-specific bias, high T and low C ratio in mitogenome was also conserved in Florometra sp. accounting for 26.33% A, 47.03% T, 11.41% C, and 15.24% G. Phylogenetic distance was estimated using the concatenated set of whole 13 PCGs of Florometra sp. mitogenome with 28 published echinoderms mitogenomes and two Balanoglossus mitogenomes as an outgroup (). The phylogenetic analysis was performed using the maximum likelihood method, GTR + G + I model with a bootstrap of 1000 replicates. The Florometra sp. mitogenome was closely clustered with the F. serratissima mitogenome. Previously, another Florometra sp. mitogenome (Accession no. AF049132) formed a cluster with that of F. serratissima (Nam et al. Citation2020). However, the incorporation of two new mitogenomes, Florometra sp. (this study) and Stephanometra indica (MF966246) on phylogenetic analysis performed in this study resulted in a newly formed cluster with that of Stephanometra indica. Although further study should be conducted to make a reliable phylogenetic relationship with appropriate species identification, our result will be a useful genomic resource for analyzing evolutionary relationship in Crinoidea.
Figure 1. Maximum-likelihood (ML) phylogeny of 29 echinoderms (6 asteroids, 7 crinoids including Florometra sp., 5 echinoids, 5 holothuroids, and 6 ophiuroids) based on the concatenated nucleotide sequences of the entire protein-coding genes (PCGs). Two Balanoglossus mitogenomes were used as an out group. Numbers on the branches indicate ML bootstrap percentages (1000 replicates). DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated. The black arrow indicates the Florometra sp. analyzed in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT302206.
Additional information
Funding
References
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Foote M. 1999. Morphological diversity in the evolutionary radiation of Paleozoic and post-Paleozoic crinoids. Paleobiology. 25(S2):1–115.
- Hess H, Ausich WI, Brett CE, Simms MJ. 1999. Fossil crinoids. Cambridge: Cambridge University Press.
- Nam S-E, Park HS, Rhee J-S. 2020. Complete mitochondrial genome of the crinoid echinoderm, Florometra species (Echinodermata, Crinoidea). Mitochondrial DNA. 5(1):852–853.
- Rouse GW, Jermiin LS, Wilson NG, Eeckhaut I, Lanterbecq D, Oji T, Young CM, Browning T, Cisternas P, Helgen LE, et al. 2013. Fixed, free, and fixed: the fickle phylogeny of extant Crinoidea (Echinodermata) and their Permian-Triassic origin. Mol Phylogenet Evol. 66(1):161–181.
- Scouras A, Smith MJ. 2001. A novel mitochondrial gene order in the crinoid echinoderm Florometra serratissima. Mol Biol Evol. 18(1):61–73.
- Wright DF, Ausich WI, Cole SR, Peter ME, Rhenberg EC. 2017. Phylogenetic taxonomy and classification of the Crinoidea. J Paleontol. 91(4):829–846.
