Abstract
Alisma plantago-aquatica is an important medicinal plant in traditional Chinese medicine. In this paper, we have reported and characterized the complete chloroplast genome of A. plantago-aquatica. Its sequence was 167,642 bp in length, which contained a large single-copy region (LSC) of 90,005 bp, a small single-copy region (SSC) of 12,209 bp, and a pair of inverted repeat (IR) regions of 32,714 bp. The overall nucleotide composition of chloroplast genome sequence is: A (53,496 bp, 31.9%), T (54,364 bp, 32.4%), C (30,346 bp, 18.1%), G (29,436 bp, 17.6%) and the total G + C content of 35.7%. The complete chloroplast genome sequence contains 136 genes, including 91 encoding genes, 37 transfer RNA genes and 8 ribosomal RNA genes. For the phylogenetic analysis result, A. plantago-aquatica is closely related to A. gramineum of the family Alismataceae in the phylogenetic relationship in this study.
Alisma plantago-aquatica is an important medicinal plant in traditional Chinese medicine that is also as Chinese Materia Medica for approximately two thousand years in China (Shu et al. Citation2016). It belongs to the genus Alisma and the family Alismataceae, which is named Ze-Xie in Chinese. Alisma plantago-aquatica was first recorded in an herbal monograph Shennong’s Classic of Materia Medica, which was extensively used to therapeutic effects on hyperlipidemia, diabetes, prophylaxis of urolithiasis, cholestasis, chronic kidney disease and cancer (Li and Qu Citation2012; Wang et al. Citation2018). Alisma plantago-aquatica is one of the important Chinese herbs in some Southeast Asia countries, but fewer genomic data and information has been reported about this species. Here, we have reported and characterized the complete chloroplast genome of A. plantago-aquatica, which can be used to resolve the deep phylogenetic relationship of the family Alismataceae that can also be useful in the traditional Chinese medicine research in the future.
The sample of A. plantago-aquatica was collected from the market of herb near the Third Hospital of Nanchang and located at Nanchang, Jiangxi, China (28.66 N, 115.90E). The corresponding voucher herbarium specimen (No. THNC-02) was stored at the Third Hospital of Nanchang. The total genome DNA was extracted from the fresh sample- of A. plantago-aquatica using the Plant Tissues Genomic DNA Extraction Kit (TIANGEN, BJ and CN), following the protocol. Then, the chloroplast genome DNA was purified and sequenced, we used FastQC (Andrews Citation2015) to control and remove the sequences. The chloroplast genome of A. plantago-aquatica was assembled using NOVOPlasty (Dierckxsens et al. Citation2017). The chloroplast genome sequence annotation was done using Geneious (Kearse et al. Citation2012). All the genes were predicted using CPGAVAS (Liu et al. Citation2012) and corrected using NCBI Blast search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The chloroplast genome sequence of A. plantago-aquatica was submitted into NCBI database and the accession number was NK9900121.
The complete chloroplast genome of A. plantago-aquatica has a typical quadripartite structure, which is consistent with most other chloroplast genomes plant species. Its sequence was 167,642 bp in length, which contained a large single-copy region (LSC) of 90,005 bp, a small single-copy region (SSC) of 12,209 bp, and a pair of inverted repeat (IR) regions of 32,714 bp. The overall nucleotide composition of chloroplast genome sequence is: A (53,496 bp, 31.9%), T (54,364 bp, 32.4%), C (30,346 bp, 18.1%), G (29,436 bp, 17.6%) and the total G + C content of 35.7%. However, the complete chloroplast genome sequence contains 136 genes, including 91 encoding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. The IR region harbors 9 protein-encoding genes (rpl2, rpl23, ycf2, ndhB, rps7, rps12, ycf1, rps15 and ndhH), 7 tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnIGAU, trnA-UGC, trnR-ACG and trnN-GUU), and all the 4 rRNA genes (rrn16, rrn23, rrn4.5 and rrn5).
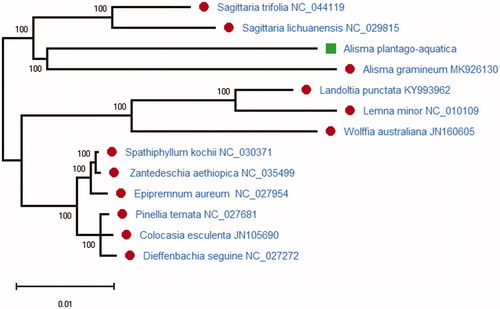
The phylogenetic relationships of A. plantago-aquatica was reconstructed based on the complete chloroplast genome of 12 plant species using the maximum-likelihood (ML) and Bayesian inference (BI) methods implemented on the MEGA X (Kumar et al. Citation2018). MEGA X was used for constructing the ML phylogenetic trees. The ML phylogenetic tree was inferred with strong support and the bootstrap values from 2000 replicates were used at all the nodes. The ML tree was drawn and edited by MEGA X. According to the phylogenetic analysis result, A. plantago-aquatica is closely related to A. gramineum of the family Alismataceae in the phylogenetic relationship (). The complete chloroplast genome of A. plantago-aquatica can be used to resolve the deep phylogenetic relationship of the family Alismataceae, which can also be useful in the traditional Chinese medicine research in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li Q, Qu H. 2012. Study on the hypoglycemic activities and metabolism of alcohol extract of Alismatis rhizoma. Fitoterapia. 83(6):1046–1053.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715–2164.
- Shu Z, Pu J, Chen L, Zhang Y, Rahman K, Qin L, Zheng C. 2016. Alisma orientale: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. Am J Chin Med. 44(02):227–251.
- Wang J, Li H, Wang X, Shen T, Wang S, Ren D. 2018. Alisol b-23-acetate, a tetracyclic triterpenoid isolated from Alisma orientale, induces apoptosis in human lung cancer cells via the mitochondrial pathway. Biochem Biophys Res Commun. 505(4):1015–1021.