Abstract
Here, we report the complete mitochondrial genome of the marine nemertean Cephalothrix species collected from the coastal region of South Korea. The mitochondrial genome of Cephalothrix sp. South Korea (SK) stain is 16,396 bp contains 13 protein-coding genes (PCGs) along with 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and an AT-rich noncoding region, arranged identically to those of Palaeonemertea. The nucleotide composition in the Cephalothrix sp. (SK) mitogenome is highly biased toward A and T as total AT content is 76.3% across the genome. A phylogenetic analysis of Nemertean species indicates that Cephalothrix sp. (SK) mitogenome shows close relationships to those of Palaeonemertea (Cephalothrix sp. and C. hongkongiensis collected from China).
The unsegmented, bilaterian, vermiform ribbon worms, nemertean are primarily marine invertebrates, ranged from less than 1 cm to several meters in body length (Gibson Citation1995; Brusca and Brusca Citation2003). Necessary in the development of accurate species description and exploration of species diversity with phylogenetic relationships has been highlighted in nemertean, while nemertean taxonomy is complex due to relatively simple biology, a few distinguishable external morphological characteristics, and difficulty of collecting preserved specimen, reports on many unrecognized cryptic species, and unclear information on their geographic distributions (Gibson Citation1995; Thollesson and Norenburg Citation2003; Strand and Sundberg Citation2005; Andrade et al. Citation2012; Sundberg et al. Citation2016). Thus, research on the phylogenetic relationship and evolutionary history in nemertean has focused on the combined diagnostic approach with external characters and genomic resource-based analysis of molecular phylogeny and genetic diversity (Chen et al. Citation2010; Andrade et al. Citation2014; Kvist et al. Citation2014; Luo et al. Citation2018). The genus Cephalothrix comprises more than 30 described species with several cryptic species and a recent study suggested that the diversity of European Cephalothrix is higher than expected based on morphological identification (Sagorny et al. Citation2019).
To date, more than 19 complete or near-complete nemertean mitogenome sequences are registered in GenBank, including two complete Cephalothrix mitogenomes (Cephalothrix sp. and C. hongkongiensis the littoral zone in Qingdao, China) (Chen et al. Citation2011) and an incomplete C. rufifrons mitogenome (Turbeville and Smith Citation2007). In this study, an individual Cephalothrix sp. was collected from Byeonsanbando (35°41′16.8″N 126°31′56.0″E) in May 2017. Total DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen Inc., Valencia, CA), following the manufacturer’s protocol. The remaining specimen is accessioned in the Research Institute of Basic Sciences of Incheon National University (Incheon, South Korea) with a specimen ID Nemertean003–2017. TruSeq DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) was used for sequencing on an Illumina HiSeq sequencer. Assembly was conducted in CLC Assembly Cell package (version 4.2.1) using the CLC de novo assemble algorithm. Additional PCR procedures were conducted to confirm the nucleotide sequence of the AT-rich noncoding region. MITOS 2 was employed to annotate (Bernt et al. Citation2013) and detailed annotation was conducted with NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
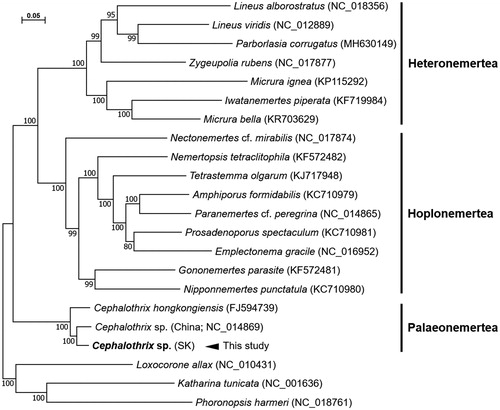
The mitochondrial genome of Cephalothrix sp. (SK) is 16,396 bp long (Accession no. MT302207). We recovered 13 PCGs, 22 tRNAs, 2 rRNAs, and an AT-rich noncoding region. All the 37 genes are predicted to be transcribed in the same direction except for trnP and trnT as shown in Cephalothrix sp. (China) and C. hongkongiensis (Chen et al. Citation2011). Approximately 89%–94% nucleotide sequence identity was observed between the 13PCGs of Cephalothrix sp. (SK) and Cephalothrix sp. (China). Quite high AT content (76.3%; A 28.5%, T 47.8%, G 14.1%, C 9.6%) was measured across the genome as shown in the genus Cephalothrix. The nucleotide composition in the Cephalothrix sp. (SK) mitogenome is highly biased toward A and T as total AT content is 76.3% across the genome. Phylogenetic analysis based on the concatenated set of 13 PCGs’ sequences of the 18 nemertean species showed that the Cephalothrix sp. (SK) mitogenome is closely related to the genus Cephalothrix ().
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT302207.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for the content and writing of this manuscript.
Additional information
Funding
References
- Andrade SCS, Montenegro H, Strand M, Schwartz ML, Kajihara H, Norenburg JL, Turbeville JM, Sundberg P, Giribet G. 2014. A transcriptomic approach to ribbon worm systematics (Nemertea): resolving the Pilidiophora problem. Mol Biol Evol. 31(12):3206–3215.
- Andrade SCS, Strand M, Schwartz M, Chen H, Kajihara H, von Döhren J, Sun S, Junoy J, Thiel M, Norenburg JL, et al. 2012. Disentangling ribbon worm relationships: multi-locus analysis supports traditional classification of the phylum Nemertea. Cladistics. 28(2):119–141.
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Brusca RC, Brusca GJ. 2003. Invertebrates. 2nd ed. Sunderland (MA): Sinauer Associates, Inc.
- Chen HX, Strand M, Norenburg JL, Sun S, Kajihara H, Chernyshev AV, Maslakova SA, Sundberg P. 2010. Statistical parsimony networks and species assemblages in Cephalotrichid nemerteans (Nemertea). PLoS One. 5(9):e12885.
- Chen HX, Sundberg P, Wu HY, Sun SC. 2011. The mitochondrial genomes of two nemerteans, Cephalothrix sp. (Nemertea: Palaeonemertea) and Paranemertes cf. peregrina (Nemertea: Hoplonemertea). Mol Biol Rep. 38(7):4509–4525.
- Gibson R. 1995. Nemertean genera and species of the world: an annotated checklist of original names and description citations, synonyms, current taxonomic status, habitats and recorded zoogeographic distribution. J Nat Hist. 29(2):271–561.
- Kvist S, Laumer CE, Junoy J, Giribet G. 2014. New insights into the phylogeny, systematics and DNA barcoding of Nemertea. Invert Systematics. 28(3):287–309.
- Luo YJ, Kanda M, Koyanagi R, Hisata K, Akiyama T, Sakamoto H, Sakamoto T, Satoh N. 2018. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat Ecol Evol. 2(1):141–151.
- Sagorny C, Wesseler C, Krämer D, von Döhren J. 2019. Assessing the diversity and distribution of Cephalothrix species (Nemertea: Palaeonemertea) in European waters by comparing different species delimitation methods. J Zool Syst Evol Res. 57(3):497–519.
- Strand M, Sundberg P. 2005. Delimiting species in the hoplonemertean genus Tetrastemma (phylum Nemertea): morphology is not concordant with phylogeny as evidenced from mtDNA sequences. Biol J Linn Soc. 86(2):201–212.
- Sundberg P, Andrade SCS, Bartolomaeus T, Beckers P, von Döhren J, Krämer D, Gibson R, Giribet G, Herrera-Bachiller A, Junoy J, et al. 2016. The future of nemertean taxonomy (phylum Nemertea)–a proposal. Zool Scr. 45(6):579–582.
- Thollesson M, Norenburg JL. 2003. Ribbon worm relationships: a phylogeny of the phylum Nemertea. Proc R Soc Lond B. 270(1513):407–415.
- Turbeville JM, Smith DM. 2007. The partial mitochondrial genome of the Cephalothrix rufifrons (Nemertea, Palaeonemertea): characterization and implications for the phylogenetic position of Nemertea. Mol Phylogenet Evol. 43(3):1056–1065.