Abstract
Syringa reticulata subsp. pekinensis (Ruprecht) P. S. Green & M. C. Chang (Oleaceae: Syringeae) is horticultural woody plant. To grasp a better comprehension on the molecular basis of S. reticulata subsp. pekinensis chloroplast, a complete characterization of the 155,819 bp chloroplast genome sequences was performed. This plastome encompassed 87,055 bp large single-copy region (LSC) and 40,064 bp small single-copy region (SSC), sandwiched by a pair of 14,350 bp inverted repeats (IRs). The GC content of the chloroplast genome was 38.06%. Moreover, a total of 131 functional genes were annotated, including 87 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The NJ phylogenetic tree based on 40 chloroplast genome sequences suggested that Syringa spp. were closely related to Ligustrum spp.
Syringa reticulata subsp. pekinensis (Ruprecht) P. S. Green & M. C. Chang (Oleaceae: Syringeae), a shrub or small tree, is a popular horticultural plant. Wild individuals of S. reticulata subsp. pekinensis grow mainly in slopes, valleys, along gullies, 600–2400 m above sea level. It usually cultivated as an ornamental in northern China (Chang et al. Citation1996). The chloroplast genome has been particularly useful for studying the maternal evolutionary history of angiosperms for its matrilinear inheritance without recombination (Nock et al. Citation2019). However, the sequence of the complete plastome of S. reticulata subsp. pekinensis. Here, the complete chloroplast genome (Genbank accession number: MN901632) was obtained based on next-generation sequencing (NGS) technologies and the phylogenetic analysis of S. reticulata subsp. pekinensis was carried out accordingly.
In this study, the samples of S. reticulata subsp. pekinensis were collected from Xining Botanical Garden, Xining City, Qinghai Province, China (36.62°N, 101.75°E). The experiment and analysis scheme refers to Wang et al. (Citation2019). The genome was extracted from the dried, young leaves (about 0.4 g) with a modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen (Specimen Accession number: WangJL2019233) was kept in Herbarium of the Northwest Institute of Plateau Biology, Chinese Academy of Sciences (HNWP). Genome sequencing was performed using the Illumina HiSeq Platform (Illumina, San Diego, CA) at Genepioneer Biotechnologies Inc., Nanjing, China. Approximately 6.78 GB of clean data were yielded. The trimmed reads were mainly assembled by CpGAVAS (Liu et al. Citation2012).
The complete chloroplast genome of S. reticulata subsp. pekinensis is 155,819 bp in length with a typical quadripartite structure, containing a pair of inverted repeated (IR) regions of 14,350 bp, a large single copy (LSC) region of 87,055 bp, and a small single copy (SSC) region of 40,064 bp. The two IRs are separated by the LSC and the SSC. The GC content of the complete chloroplast genome was 38.06%. A total of 131 functional genes were annotated, including 8 rRNA genes, 36 tRNA genes, and 87 protein-coding genes. The rRNA genes, tRNA genes, and protein-coding genes account for 6.11%, 27.48%, and 66.41% of all annotated genes, respectively.
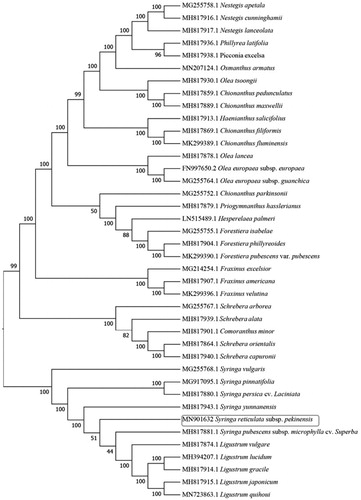
Phylogenetic relationships of S. reticulata subsp. pekinensis with 39 other species of Oleaceae were resolved by means of neighbor-joining (NJ). Alignment was conducted using MAFFT (Katoh and Standley Citation2013; online version: https://mafft.cbrc.jp/alignment/server/). The NJ was built using MEGA version 7 (MEGA, Philadelphia, PA) (Kumar et al. Citation2016) with bootstrap set to 1000. The NJ phylogenetic tree suggested that Syringa spp. were closely related to Ligustrum spp. ().
Data availability statement
The data that support the findings of this study are openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MN901632.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chang MC, Qui LQ, Wei Z, Green PS. 1996. Oleaceae. In: Wu Z, Raven PH, editors. Flora of China. Beijing (China): Science Press and Missouri Botanical Garden; p. 272–319.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Nock CJ, Hardner CM, Montenegro JD, Termizi AAA, Hayashi S, Playford J, Edwards D, Batley J. 2019. Wild origins of macadamia domestication identified through intraspecific chloroplast genome sequencing. Front Plant Sci. 10:334.
- Wang J, Cao Q, Wang K, Xing R, Wang L, Zhou D. 2019. Characterization of the complete chloroplast genome of Pterygocalyx volubilis (Gentianaceae). Mitochondrial DNA Part B. 4(2):2579–2580.