Abstract
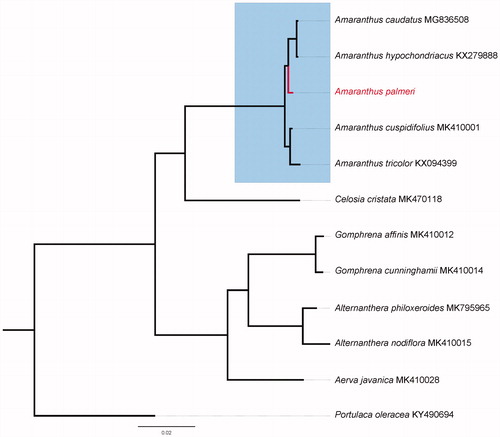
The complete chloroplast genome of Palmer Amaranth Amaranthus palmeri was sequenced and assembled in this study. The genome is 150,721 bp in length and contained 129 encoded genes in total, including 84 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. The phylogenetic tree based on 12 chloroplast genomes in Amaranthaceae support that A. palmeri is sister to A. hypochondriacus and A. caudatus.
Amaranthus palmeri S. Watson, a dioecious summer annual forb, is originally native to the southwestern United States and northwestern Mexico. For the past decades, A. palmeri dramatically expanded its distribution throughout the United States and has become a hard-to-control weed that severely compromises crop yields in agriculture production systems (Ward et al. Citation2013; Davis et al. Citation2015). Moreover, this weedy species with invasive behavior has been transported recently to many other countries in North America, South America, Asia, Africa, and Europe through international grain trades (Kistner and Hatfield Citation2018). No complete organelle molecular data have been reported for this species to our knowledge. In this study, we assembled and characterized the first A. palmeri complete chloroplast genome. It will provide potential genetic resources for DNA barcoding marker development and quarantine inspection.
Leaves of A. palmeri were collected from Riverside, California, USA (GPS: 33°50′12.1″N, 117°07′25.0″W, Voucher YW201808009, deposited at Chenshan Botanical Garden). DNA was extracted from its silica dried leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The plastome sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). In total, ca. 20.9 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. Following Liu et al (Citation2017, Citation2018), aligning, assembly, and annotation were conducted by CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and GENEIOUS v 11.0.5 (Biomatters Ltd, Auckland, New Zealand).
The full length of A. palmeri chloroplast sequence (GenBank Accession No. MT331487) is 150,721 bp. It comprised of a large single-copy region (LSC with 83,997 bp), a small single-copy region (SSC with 18,030 bp), and two inverted repeat regions (IR with 24,347 bp). The GC content of A. palmeri chloroplast genome was 36.6%. A total of 127 genes were contained in the genome (84 protein-coding genes, 8 rRNA genes, and 37 tRNA genes). Nineteen genes had two copies, which were comprised of 7 PCG genes (rps12, rpl2, rpl23, ndhB, rps7, ycf2, rps19), 8 tRNA genes (trnH-GUG, trnl-CAU, trnL-CAA, trnv-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU), and all 4 rRNA species (rrn16, rrn23, rrn4.5, rrn5). In the genome, 9 protein-coding genes (rps16, atpF, rpoc1, petB, petD, rpl16, ndhB, ndhA, rpl2) had one intron, and 4 protein-coding gene contained two introns.
Eleven species with available chloroplast genomes were selected to study the phylogenetic placement of A. palmeri in Amaranthaceae (). The sequence alignment was conducted using MAFFT v7.3 (Katoh and Standley Citation2013). The phylogenetic tree was constructed using IQTREE v1.6.7 (Nguyen et al. Citation2015), with the best selected GTR + F+R3 model and 5000 bootstrap replicates. The result indicated that A. palmeri is sister to A. hypochondriacus and A. caudatus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The DNA matrix and phylogenetic tree that support the findings of this study are openly available in Github at https://github.com/andresqi/Amaranthus-palmeri-chloroplast-genome
Additional information
Funding
References
- Davis AS, Schutte BJ, Hager AG, Young BG. 2015. Palmer amaranth (Amaranthus palmeri) damage niche in Illinois soybean is seed limited. Weed Sci. 63(3):658–668.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kistner EJ, Hatfield JL. 2018. Potential geographic distribution of Palmer amaranth under current and future climates. Agric Environ Lett . 3(1):170044.
- Liu L, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu C. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of fagales. Front Plant Sci. 8:968
- Liu LX, Li P, Zhang HW, Worth JRP. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res Jpn. 23(5):318–323.
- Nguyen L, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Ward SM, Webster TM, Steckel LE. 2013. Palmer amaranth (Amaranthus palmeri): a review. Weed Technol. 27(1):12–27.