Abstract
The genus Mammillaria occupies diverse habitats and exhibits diverse growth patterns and a large range of morphologies. Most of the species of this genus are used as ornamental plants and are subject to mass habitat loss. Due to these factors, they are being submitted to selective pressure that might affect conservational efforts and management plans. We obtained the 133 gene chloroplast genome as part of the project of sequencing the complete genome of pincushion cactus, including 88 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The phylogenetic tree indicates the pincushion cactus is a sister species of M. supertexta and M. huitzilopochtli.
The Mammillaria genus is comprised of small succulent plants, which are highly diverse in growth habits and morphologies, making it the most species-rich genus within the Cactaceae family and among the top five species-rich genus in Mexican vascular plants (Villaseñor Citation2016). Among its 166 species distributed in the Americas, 150 species are endemic to Mexico, with many species having narrow distributions (Hernandez and Godinez Citation1994). This species requires more comprehensive and robust conservation strategies than widely distributed taxa; like the pincushion cactus (Mammillaria haageana subsp. san-angelensis), whose distribution is restricted to the Ecological Reserve of Pedregal de San Ángel (REPSA) in Mexico City (Arias Citation2009; Valverde and Chávez Citation2009). This makes the species an ideal model to understand ecological and evolutionary implications. Here we present the chloroplast genome of M. haageana subsp. san-angelensis as an initial approach toward developing genomic tools to better understand M. haageana and its close relatives.
We sampled an individual of the pincushion cactus (JardínBotánico/UNAM = C-IC-02-03) from the live collection of the University Botanical Garden (19.3206° N, 99.1944° W). Total genomic DNA was extracted using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. High-throughput DNA sequencing was conducted using the NovaSeq6000 System with S1 FlowCell (Illumina Inc., San Diego, CA) 2 × 150 bp. After base quality control using Trimmomatic v0.32 (Bolger et al. Citation2014), the remaining high-quality reads were used to assemble the chloroplast genome by Novoplasty 3.7.1 (Dierckxsens et al., Citation2016) using M. supertexta chloroplast genome reference (Solórzano et al. Citation2019) and rpl16 gene (Genbank accession number: AY545323.1) as seed, following the software specifications (Dierckxsens et al., Citation2016). The plastome was annotated using GeSeq (Tillich et al., Citation2017). The accurate gene boundaries were confirmed by alignment with other chloroplast genes of Cactaceae using MAFFT v7.311 (Katoh et al., Citation2019). We generated a maximum-likelihood (ML) tree through IQTREE (Nguyen et al. Citation2015) with 1000 bootstrap alignments and SH-aLRT and Bayes tests with 1000 replicates based on 11 complete chloroplast sequences aligned with Dionaea muscipula as outgroup.
The complete chloroplast genome of M. haageana subsp. sanangelensis was determined to be 115,386 bp in length, similar to those previously reported by Solórzano et al. 2019 for another Mammillaria species. The M. haageana subsp. san-angelensis genome has 133 genes, including 88 protein-coding genes, 8 rRNA genes, and 37 tRNA genes.
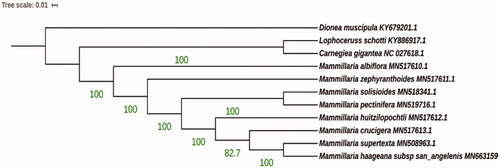
The ML analysis reveals that M. haageana subsp. san-angelensis is a sister to M. supertexta forming monophyletic group closely related to M. crucigera and M. huitzilopochtli ().
Figure 1. Molecular phylogeny of M. haageana subsp. san-angelensis constructed by Maximum likelihood method. Mammilllaria haageana subsp. san-angelensis has been considered a subspecies for more than 30 years, but our initial results may indicate that it could be elevated to species status. Further studies including complete chloroplast phylogenomics and morphology will be useful to elucidate the Mammillaria phylogenomics.
Acknowledgements
We thank the Programa de Becas Posdoctorales UNAM and Posgrado en Ciencias Biológicas UNAM for SH-A fellowship to conduct this postdoctoral research. We thank Jerónimo Reyes-Santiago for providing the sequenced plant, and Cristian Cervantes-Salgado for the DNA extraction. We thank Luis Zambrano and Zenon Cano for their support as part of the REPSA project IV200117. Zoë Frezques for proof-reading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Arias S. 2009. La familia Cactaceae. Biodiversidad del ecosistema del Pedregal de San Ángel. México (DF): Universidad Nacional Autónoma de México. p. 135–140.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hernandez HM, Godinez H. 1994. Contribución al conocimiento de las cactáceas mexicanas amenazadas. Acta Bot Mex. 26:33–52.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32:268–274.
- Solórzano S, Chincoya DA, Sanchez-Flores A, Estrada K, Díaz-Velásquez CE, González-Rodríguez A, Vaca-Paniagua F, Dávila P, Arias S. 2019. De novo assembly discovered novel structures in genome of plastids and revealed divergent inverted repeats in Mammillaria (Cactaceae, Caryophyllales). Plants. 8(10):392.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Valverde T, Chávez VM. 2009. Mammillaria (Cactaceae) como indicadora del estado de conservación del ecosistema. Biodiversidad del ecosistema del Pedregal de San Ángel. México (DF): Universidad Nacional Autónoma de México. p. 497–507.
- Villaseñor JL. 2016. Checklist of the native vascular plants of Mexico. Revista Mexicana de Biodiversidad. 87(3):559–902.
