Abstract
Betula hainanensis (Betulaceae) is a new reported species of the genus Betula, which is distributed in tropical, montane rain forests and evergreen broad-leaved forests in the Hainan Island, China. The complete chloroplast genome of B. hainanensis is a typical quadripartite cycle with 161,189 bp length, including a large single-copy region (LSC, 89,841 bp), a small single-copy region (SSC, 19,318 bp), and a pair of 26,015 bp inverted repeat regions (IRs). The circular genome contained 129 genes, including 84 protein-coding genes, 8 ribosomal RNA genes, and 37 tRNA genes. The overall GC content of the circular genome was 35.9%. A maximum likelihood (ML) tree showed a close relationship between B. hainanensis and B. alnoides.
Betula hainanensis J. Zeng, B.Q. Ren, J.Y. Zhu & Z.D. Chen, was reported as a new species of the genus Betula in 2014; it is distributed in tropical, montane rain forests and evergreen broad-leaved forests at altitude of above 700 m in Hainan Island, China (Zeng et al. Citation2014). Its morphology and phenology traits and the gene ITS sequences show differences from B. alnoides, B. luminifera and B. fujianensis (unpublished). As B. hainanensis is reported recently, its phylogenetic relationships with other species of genus Betula are not clear. The paternal inheritance characteristic of chloroplast plasmid in angiosperms plants makes it an excellent tool for phylogenetic analysis. Here, we obtained the complete chloroplast genome sequence of B. hainanensis, which will help us determine its phylogenetic position.
Genomic DNA of B. hainanensis was extracted from silica gel-dried leaves collected from Jianfeng Mountain (Hainan, China; 18°45′04″N, 108°51′50″E) with a modified cetyl trimethyl ammonium bromide (CTAB) method (Zeng et al. Citation2002). The leaves specimen and its DNA are deposited at the Herbarium of Research Institute of Tropical Forestry, Chinese Academy of Forestry (Guangzhou, Guangdong, China), and the specimen accession number is BH-RITF-GYZ-JFM-103. A genomic shotgun library with an insert size of 350 bp was prepared, and then was sequenced on Illumina Hiseq4000 Platform (Illumina, San Diego, CA). About twenty million high-quality reads were mapped to B. alnoides chloroplast genome (Yin et al. Citation2019) with Geneious (Kearse et al. Citation2012), and the complete chloroplast genome was assembled with SPAdes (Bankevich et al. Citation2012). The assembled chloroplast genome was then annotated on Plann and corrected with Sequin (Huang and Cronk Citation2015). The complete chloroplast genome sequence of B. hainanensis with genes annotated was submitted to the GenBank (Accession number MT013361).
The complete chloroplast genome of B. hainanensis was circular in shape with 161,189 bp length, which possessed a large single-copy region (LSC, 89,841 bp), a small single-copy region (SSC, 19,318 bp), and a pair of 26,015 bp inverted repeat regions (IRs). The circular genome contained 129 genes, including 84 protein-coding genes, 8 ribosomal RNA genes and 37 tRNA genes. Most of the genes were in a single copy, while 16 genes occurred in double copies, including four rRNA genes (4.5S, 5S, 16S, and 23S rRNA), seven tRNA genes, and five PCG genes. The base composition of the chloroplast genome was 31.5% A, 32.5% T, 18.3% C and 17.6% G. The overall GC content of the circular genome was 35.9%, while the corresponding values of the LSC, SSC and IR regions were 33.5%, 29.3% and 42.5%, respectively.
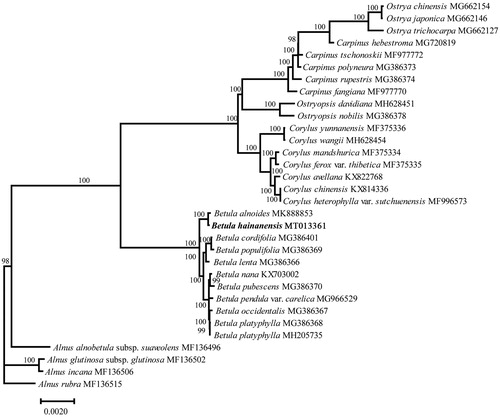
To investigate the phylogenetic relationships of Betula species, we rebuild a maximum likelihood (ML) tree with the complete chloroplast genomes of other 9 Betula species and 21 species from related genera. MAFFT (Katoh and Standley Citation2013) was used to align the chloroplast genome sequences. Model Finder (Kalyaanamoorthy et al. Citation2017) was used to select model on basis of the Bayesian information criterion (BIC), and IQ-TREE to construct the ML tree with 1000 bootstrap replicates (Nguyen et al. Citation2015). The tree showed a close relationship between B. hainanensis and B. alnoides (). The report of complete chloroplast genome of B. hainanensis will help us get insight into studies on conservation and evolutionary histories for this species.
Figure 1. Phylogenetic tree based on chloroplast genomes using the ML method. Ultrafast bootstrap values are shown above the nodes, with 1000 bootstrap replicates. The chloroplast genome sequences of 31 individuals of 30 species except for Betula hainanensis were downloaded from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov) with the accession number is MT013361, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Yin MY, Guo JJ, Zeng J. 2019. Complete chloroplast genome sequence of Betula alnoides (Betulaceae). Mitochondr DNA B. 4(2):2409–2410.
- Zeng J, Ren BQ, Zhu JY, Chen ZD. 2014. Betula hainanensis (Betulaster, Betulaceae), a new species from Hainan Island, China. Ann Bot Fennici. 51(6):399–402.
- Zeng J, Zou YP, Bai JY, Zheng HS. 2002. Preparation of total DNA from ‘recalcitrant plant taxa. Acta Bot Sin. 44:694–697.
