Abstract
Rosa laevigata var. leiocarpus has excellent edible, ornamental and medicinal value in China. In this study, we presented the complete chloroplast genome of R. laevigata var. leiocarpus. The whole chloroplast genome is 156,373 bp in size, consisting of a pair of inverted repeats (IR 26,047 bp), a large single-copy region (LSC 85,494 bp) and a small single-copy region (SSC 18,785 bp). The R. laevigata var. leiocarpus chloroplast genome encodes 139 annotated known unique genes including 92 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on chloroplast genomes of 18 plant species reveals that R. laevigata var. leiocarpus was clustered and closest with Rosa roxburghii and also closer to other Rosa species in the evolutionary relationship.
Rosa laevigata var. leiocarpus is a variant of R. laevigata found in Boluo county, Guangdong province, China, was firstly reported by Wang and Chen in 1995 (Wang and Chen 1995) (CitationYingqiang W,Pangyu C). Rosa laevigata var. leiocarpus and R. laevigata have good edible, ornamental and medicinal value. The most significant different variation of R. laevigata var. leiocarpus is that the leaves are thinner and the fruit is smooth and spiny, andnd this particular morphological feature makes it easier to be collected and prepared (Hua et al. Citation2010). Therefore, we reported the complete chloroplast genome of R. laevigata var. leiocarpus and explored the phylogenetic relationship with other plant species in Rosaceae family, which would be helpful for its evolution and genetics research.
A wild individual of R. laevigata var. leiocarpus was sampled from Xia lin, Dongyuan village, Heyuan City, Guangdong province, China (N23°97′6.65″, E114°71′1.47″). The voucher specimen was deposited in the Herbarium of South China Agricultural University (CANT) under the accession number of CDBI: LLS0006-2. A genome library was constructed using TruSeq DNA sample preparation kit (Illumina, USA) with insertion size of 300 bp and sequenced on Illumina HiSeq Nova platform (guangzhou gerui biotechnology). The output data was a 6GB raw data, 150 bp double-ended read. Clean data were obtained by quality filtering of the raw data, and then the chloroplast genome assembly of plants was carried out by the software GetOrganelle (Jin et al. Citation2018). The assembled cp genome was annotated with Geseq (Tillich et al. Citation2017) and compared manually with the complete cp genome for R. laevigata var. leiocarpus (Genbank accession number: MT150578).
The complete cp genome of R. laevigata var. leiocarpus was determined. The whole chloroplast genome is 156,373 bp in size, consisting of a pair of inverted repeats (IR 26,047 bp), a large single-copy region (LSC 85,494bp) and a small single-copy region (SSC 18,785bp). The R. laevigata var. leiocarpus chloroplast genome encodes 139 annotated known unique genes including 92 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. This structure is identical to those of other species in Rosaceae.
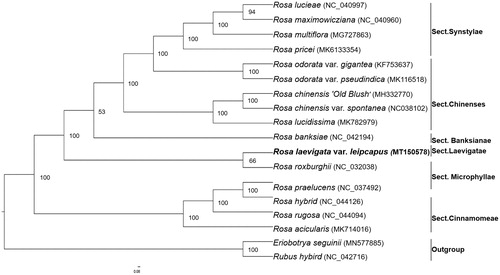
We selected other 17 plants species chloroplast genomes from GenBank to assess the relationship of R. laevigata var. leiocarpus. The phylogenetic tree was reconstructed using maximum-likelihood (ML) methods and performed using IQ-TREE (Nguyen et al., Citation2015), with the bootstrap value calculated using 1000 replicates to assess node support. The final tree was edited using the iTOL v4 (Ivica and Peer Citation2019). As shown in the phylogenetic tree result (), the chloroplast genome of R. laevigata var. leiocarpus was clustered and closest with R. roxburghii, the genus Laevigatae was monophyletic and clustered as the sister group to R. Sect. Banksianae. As the data of R. laevigata were not released online, no comparison was conducted. Further study in the future will give a better understanding of its system status. This discovery provides valuable genomic resources for the future development of medicinal plants and phylogeny of Rosaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hua L, Hong P, Fen C, Hua H. 2010. Comparative study on the quality of Rosalaevigata var. leiocarpus and Rosa laevigata Michx. J Guangdong Pharm Coll. 26(04):345–347.
- Ivica L, Peer B. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256-w259
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.
- Nguyen L-T, Schmidt H A, Von Haeseler A, Minh B Q. 2015. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution. 32(1):268–274. doi:10.1093/molbev/msu300.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yingqiang W, Pangyu C. 1995. New Taxa of Guangdong Plants.Journal of Tropical and Subtropical Botany. 3(1):29–33.