Abstract
The complete chloroplast genome of Chrysosplenium macrophyllum Oliv. and Chrysosplenium flagelliferum Fr. Schmidt. were reported in this study. The chloroplast genomes were 152,837 bp for C. macrophyllum and 151,679 bp for C. flagelliferum. LSC and SSC of 83,584 bp and 17,265 bp were separated by two IRs of 25,994 bp each in C. macrophyllum. While C. flagelliferum contained IRs of 25,973 bp, LSC of 82,772 bp and SSC of 16,961 bp, for a total 151,679 bp length. The chloroplast genome of Chrysosplenium macrophyllum contains 130 genes, including 85 protein-coding genes (78 PCG species), 8 ribosomal RNA genes (4 rRNA species), 37 transfer RNA genes (30 tRNA species). And the chloroplast genome of Chrysosplenium flagelliferum contains 130 genes, including 85 protein-coding genes (78 PCG species), 8 ribosomal RNA genes (4 rRNA species), 37 transfer RNA genes (30 tRNA species).
Keywords:
Chrysosplenium L. (Saxifragaceae) comprises about 65-70 species in the worldwide, mainly distributed in the northern hemisphere (Hara Citation1957; Soltis et al. Citation2001; Lan et al. Citation2018; Liao et al. Citation2019). They grow in moist, shaded valleys and mountain slopes (Kim et al. Citation2018). According to leaf arrangement, alternate or opposite, Chrysosplenium is divided into two subgenera: Alternifolia Franchet and Oppositifolia Franchet (Hara Citation1957). In this study, we report and characterize two chloroplasts genomes from two species: Chrysosplenium macrophyllum from subgenus Alternifolia and Chrysosplenium flagelliferum from subgenus Oppositifolia. Using these data, we reconstruct the phylogenetic tree of this genus to reveal the relationship and provide useful information for further study of Chrysosplenium.
The materials of C. macrophyllum and C. flagelliferum were collected from Hubei, China (BD2017030507344, N 30°44’04”, E 110°15’26”) and Tochigi-ken, Japan (RG2019032810002, N 36° 45′32″, E 139°36′01″). The voucher specimens were deposited at the Herbarium of South-Central University for Nationalities (HSN). The chloroplast DNA of C. macrophyllum was extracted by a high salt method (Shi et al. Citation2012) and sequenced using the PacBio II Platform at Frasergen (Wuhan China). The genome assembly was conducted using Canu-v1.5 (Koren et al. Citation2017). To discard the sequence of the nucleus, we aligned the contigs to the whole chloroplast data from NCBI. Then the draft chloroplast genome was polished with Arrow (SMRT link v6.0.0). However, the complete genomic DNA of C. flagelliferum was extracted using CTAB method and sequenced using the Illuminaplatform at Novogene Company (Beijing, China). After filtered the low-quality data and adaptors, the obtained clean data were aligned to C. macrophyllum with bwa-0.7.12. The aligned reads were then assembled with ABYSS-2.0.2 after the best Kmer was chosen with kmergenie. Then, connected the overlap and scaffolding again by SSPACE_Standard_v3.0. Finally, the gaps were filled by Sanger.
The complete chloroplast genome of C. macrophyllum (MK973001) was 152,837 bp in length and composed of two inverted repeats (IRs) of 25,995 bp which divide LSC of 83,583 bp and SSC of 17,264 bp, the average GC content was 37.46%. On the other hand, the complete chloroplast genome size of C. flagelliferum (MN729584) was 151,679 bp in length, the average GC content was 37.43%. Separating LSC of 82,771 bp and SSC of 16,960 bp, a pair of IRs was 25,974 bp long in each. The chloroplast genome of Chrysosplenium macrophyllum contains 130 genes, including 85 protein-coding genes, 8 ribosomal RNA genes, 37 transfer RNA genes. The chloroplast genome of Chrysosplenium flagelliferum contains 130 genes, including 85 protein-coding genes, 8 ribosomal RNA genes, 37 transfer RNA genes.
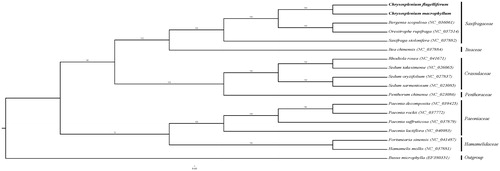
Phylogenetic analysis was performed using whole chloroplast coding sequences of C. macrophyllum and C. flagelliferum, combining with three species of Saxifragaceae, four species of Crassulaceae, four species of Paeoniaceae, two species of Hamamelidaceae, Iteaceae, Penthoraceae, and Buxus microphylla as outgroup. The phylogenetic relationships were reconstructed by means of maximum-likelihood (ML) with the model of GTR + F+R3. Based on the phylogenetic tree, we can see that C. macrophyllum and C. flagelliferum have a close relationship with Bergenia scopulosa ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hara H. 1957. Synopsis of the genus Chrysosplenium (Saxifragaceae). J Fac Sci Univ Tokyo. 7:1–90.
- Kim YI, Lee JH, Kim YD, 2018. The complete chloroplast genome of a Korean endemic plant Chrysosplenium aureobracteatum. YI, Kim YD. Kim (Saxifragaceae). Mitochondrial DNA Part B. 3(1):380–381.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736.
- Lan DQ, Liu H, Huang W, Yi LS, Zhang DD, Qin ED, Qin R. 2018. Investigation and analysis of Chrysosplenium resources in Tibetan-inhabited area. JPGR. 20:662–668.
- Liao R, Dong X, Wu ZH, Qin R, Liu H. 2019. Complete chloroplast genome sequence of Chrysosplenium sinicum and Chrysosplenium lanuginosum (Saxifragaceae). Mitochondrial DNA Part B. 4(2):2142–2143.
- Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PloS One. 7(2):e31468–e31468.
- Soltis DE, Tago-Nakazawa M, Xiang QY, Kawano S, Murata J, Wakabayashi M, Hibsch-Jetter C. 2001. Phylogenetic relationships and evolution in Chrysosplenium (Saxifragaceae) based on matK sequence data. Am J Bot. 88(5):883–893.