Abstract
Epicauta ruficeps is widely distributed in China and some countries in Southeast Asia, and plays an important role in medicine and biological control. The complete mitochondria genome of E. ruficeps was 15,813 bp in length, with 37 genes, including 13 PCGs, 22 tRNA genes (tRNAs), and two rRNA genes (rRNAs). The positions and sequences of genes were consistent with those of known Meloidae species. The nucleotide composition was highly A + T biased, accounting for ∼65% of the whole mitogenome. The complete mitogenome of E. ruficeps would help understand Meloidae evolution.
Meloidae insects’ unique life cycle, larval parasitism, adult gregariousness, chemical defense, drought tolerance and other interesting biological characteristics, make this group classification and other studies receive extensive attention (Wang et al. Citation2010). Epicauta ruficeps (Coleoptera: Meloidae) is widely distributed in China and some countries in Southeast Asia, and plays an important role in medicine and biological control (Wang et al. Citation2010). The adult is able to secrete cantharidin which has obvious anticancer and insecticidal properties (Moed et al. Citation2001; Fang et al. Citation2001; Li et al. Citation2009). In this study, we reported the complete mitochondrial genome of E. ruficeps and phylogenetic analysis for the first time (GenBank accession No: MN913338). The sample of E. ruficeps was collected from Sanming (26°13′N, 117°36′E), Fujian Province, China, in June 2019, and the specimens are deposited in Key Laboratory of Integrated Pest Management in Ecological Forests, Fujian Agriculture and Forestry University, Fuzhou, China (voucher no. ER-201906). DNA materials were extracted from tissues using TruSeq DNA sample Preparation kit (Vanzyme, China). The complete mitochondrial genomes were obtained through Illumina Hiseq 2500.
The complete mitogenome of E. ruficeps was circular in shape and 15,813 bp in length with 35.36% GC content. In total, 14,496 genes were annotated, containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs), and two ribosomal RNA (rRNAs), and 1317 nucleotides were non-coding DNA (D-loop region). The composition and arrangement of the mitogenome was comparable to the case of other Meloidae (Jie et al. Citation2016; Wu et al. Citation2018). The nucleotide composition was highly A + T biased, the A + T content of PCGs, tRNAs, and rRNAs was 64.96%, 71.62%, and 71.47%, respectively. The first genes started from trnM in the same direction with other Meloidae insects (Jie et al. Citation2016; Wu et al. Citation2018).
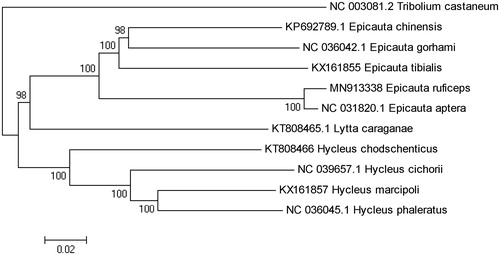
Phylogenetic analysis was constituted by 13 PCGs sequence of E. ruficeps the with the other 9 species of Meloidae, and we chose Tribolium castaneum as outgroup. The phylogenetic tree was built by using the Neighbor-Joining (NJ) method with 1000 bootstrap replicates through MEGA 7.0 (Sudhir et al. Citation2016). The result showed that E. ruficeps was closely related to E. aptera, they evolved earlier than other species of Epicauta, and the phylogenetic relationship among Meloidae was (((Epicauta) + Lytta) + Hycleus) (). In conclusion, the complete mitogenome of E. ruficeps reported in this study provided important information for the phylogeny and evolution analysis of Meloidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in “NCBI” at https://www.ncbi.nlm.nih.gov/, reference number MN913338.
Additional information
Funding
References
- Fang YL, Tan JJ, Ma WZ, Liu JP, Liu X. 2001. The natural resource and content of cantharidin in the adult of meloids in China. Acta Entomol Sinica. 44(2):192–196.
- Jie H, Lei MY, Li PM, Feng XL, Zeng DJ, Zhao GJ, Zhu JB, Zhang CL, Yu M, Huang Y, et al. 2016. The complete nucleotide sequence of the mitochondrial genome of Epicauta aptera Kaszab. Mitochondrial DNA Part B. 1(1):489–490.
- Li XF, Hou XH, Chen XS. 2009. Inhibitory effect of cantharidin from meloids on laryngeal carcinoma cell lines and gastric carcinoma cell lines. Acta Entomol Sinica. 52(9):946–951.
- Moed L, Shwayder TA, Chang MW. 2001. Cantharidin revisited: a blistering defense of an ancient medicine. Arch Dermatol. 137(10):1357–1360.
- Sudhir K, Glen S, Koichiro T. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Wang XP, Pan Z, Ren GD. 2010. The Chinese genera of Meloidae (Coleoptera: Tenebrionoidea). Entomotaxonomia. 32:45–32.
- Wu YM, Liu YY, Chen XS. 2018. The complete mitochondrial genomes of Hycleus cichorii and Hycleus phaleratus (Coleoptera: Meloidae). Mitochondrial DNA Part B. 3(1):159–160.