Abstract
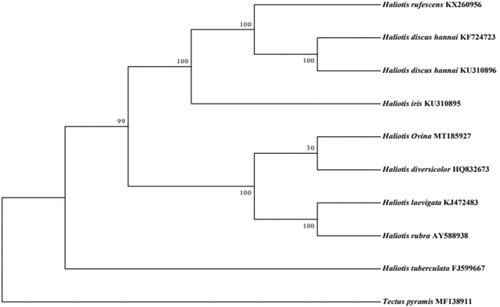
The complete mitochondrial genome of Haliotis ovina, an economically valuable abalone in China, was sequenced and analyzed. The mitogenome was 16,531 bp in length and contained 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a putative control region. The gene arrangement and orientation were identical with other Haliotis species. Phylogenetic tree was constructed based on 13 PCGs of 8 Haliotidae species, the tree revealed that H. ovina was more closely related to Haliotis diversicolor.
Abalones (family Haliotidae) are ecologically and economically important marine gastropods mollusk that are considered a great delicacy, belonging to marine gastropods and is widely distributed along the coastal waters of tropical and temperate areas (Geiger Citation1999). Haliotis ovina, which is also mainly distributed in tropical areas, has a high economic importance because it is both fished and farmed (Jarayabhand and Paphavasit Citation1996). The analysis of mitochondrial genome variation can be used to study stock structure and hybridization, and can provide useful information with implications for the management of the fishery.
The sample of H. ovina was obtained at Paracel Islands (16°28′19.71″N, 111°44′55.17″E), China. After dissection, the foot muscle was deposited in the Marine College of Hainan University (Accession number: HYXY205PSW01). The total DNA was extracted using a TIANamp Marine Animals DNA Kit (TIANGEN Biotech Co., Ltd.). The libraries were sequenced on the HiSeq (Illumina) producing total data. The quality of sequencing reads was assessed using FastQC (Andrews Citation2010). De novo assembly and sequence annotation were performed using MitoZ (Meng et al. Citation2019).
The complete mitogenome of H. ovina (GenBank Accession no. MT185927) was 16,531 bp in length with an overall 41% GC content. The mitogenome was coded for 37 genes, including 13 protein-codinggenes (PCGs), 2 rRNA, and 22 tRNA genes. The protein-coding genes included two ATP synthase subunits (ATP6, ATP8), an apocytochrome b (CYTB), three subunits of the cytochrome oxidase (COX1-3), and seven subunits of the NADH dehydrogenase (ND1-6, ND4L). All 13 PCGs were initiated by the start codon ATG, except for ND4 ND5 ND6, seven PCGs were terminated by a TAG codon, the genes in the ND3, COX1, COX2, ATP8, and ND1 cluster were terminated by a TAA codon. The 16S rRNA was 1446 bp with 61.1% AT, while 12S rRNA was 1007 bp with 59.8% AT. The 22 tRNAs were 63–73 bp long with the shortest tRNA being lysine, and the longest, tryptophan.
Genome sequences were aligned using the software Mafft version 7 (Katoh and Standley Citation2013). The phylogenetic analysis was performed using RAxML 8.2.12 software (1000 bootstrap replicates, -f a option, Stamatakis Citation2014) based on the concatenated nucleotide sequences of the 13 PCGs of 8 Haliotis species, including H. discus hannai Chinese population (Guo et al. Citation2019) and Korea population (Yang et al. Citation2015), available in GenBank, Tectus pyramis was used as an outgroup (). The phylogenetic tree revealed that 8 Haliotis species constituted three subclades, H. Ovina, H. diversicolor, H. laevigata, and H. rubra clustered in one clade; H. discus hannai (Chinese population and Korea population), H. rufescens, and H. iris clustered in another clade,; the H. tuberculata clustered in one clade. The tree revealed that H. ovina was more closely related to H. diversicolor.
Figure 1. Phylogenetic tree showing the relationship between H. ovina (GenBank: MT185927) and 7 other abalone species mitochondrial genomes. The sequences from 13 protein-coding genes were used to build the tree. Using maximum likelihood method. Numbers above the nodes indicate 1000 bootstrap values. The GenBank accession numbers are given after the species names.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, GenBank accession number MT185927.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Geiger DL. 1999. Distribution and biogeography of the recent Haliotidae (Gastropoda: Vetigastropoda) world-wide, Bollettino malacologico. Int J Malacol. 35:57–120.
- Guo Z, Ding Y, Han L, Hou X. 2019. Characterization of the complete mitochondrial genome of Pacific abalone Haliotis discus hannai. Mitochondrial DNA. 4(1):717–718.
- Jarayabhand P, Paphavasit N. 1996. A review of the culture of tropical abalone with special reference to Thailand. Aquaculture. 140(1–2):159–168.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yang EC, Nam BH, Noh SJ, Kim YO, Kim DG, Jee YJ, Park JH, Noh JH, Yoon HS. 2015. Complete mitochondrial genome of Pacific abalone (Haliotis discus hannai) from Korea. Mitochondrial DNA. 26(6):917–918.
