Abstract
In the present study, the complete mitochondrial genome of Microphysogobio tungtingensis has been amplified with 16 pairs of primers. There are 16 627 base pairs has been identified and deposited in the GenBank with accession numbers MN970213. The arrangement was similar to typical vertebrate mitochondrial, including 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs genes and a noncoding control region. The overall base composition of M. tungtingensis was G + C: 42.9%, A + T: 57.1%, apparently with a slight AT bias. Phylogenetic analysis showed that M. tungtingensis was close to M. fukiensis.
Twenty species of Microphysogobio have been found in China (Huang et al. Citation2018) while 30 species were identified in the world (Huang et al. Citation2017). Microphysogobio tungtingensis is a genus of small benthic gudgeons with 60 mm in length, which is widely distributed in the Dongting Lake and other streams. There are significant genetic divergence between Microphysogobio (Huang et al. Citation2017), further study of the taxonomic status among Microphysogobio is necessary with more species. For the above-mentioned purpose, Microphysogobio tungtingensis was obtained from the Wuyang River (N108.15, E27.04) and soaked with ethyl alcohol (95%) in Guizhou University. The total genomic DNA was extracted from skeletal muscle tissues of the fishes with the DNA extraction Kit (Chen et al. Citation2012) (Invitrogen, Carlsbad, CA).
The mitogenome had the typical vertebrate mitochondrial gene arrangement, including 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs genes, and a noncoding control region (CR) (Yang et al. Citation2016). CR of 927 bp length is located between tRNAPro and tRNAPhe. 22 tRNAs are interspersed between ribosomal RNA and protein-coding genes and range in size from 65 to 80 nucleotides.
In order to reassess the taxonomic status of Microphysogobio, 12 species were chosen. Phylogenetic analyses were constructed by the neighbor-joining (NJ) method performed on total mitogenomes (Zou et al. Citation2017) ().
Figure 1. The phylogenetic analyses investigated using N-J analysis indicated evolutionary relationships among 12 species based on total mitogenomes. Biw: Biwia, Mic: Microphysogobio, Pla: Platysmacheilus, Cyp:Cyprinidae.
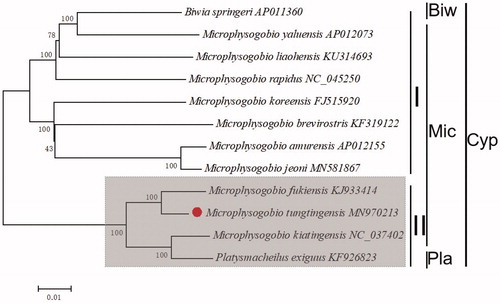
The phylogenetic tree revealed that the Microphysogobio could be divided into two clades (I and II). In group I, there were 8 species clustered together, while the group II with 4. However, the out group Biwia springeri clustal together with group, Platysmacheilus exiguus is sister to M. koreensis which were contrary to expectations. Our work confirmed that the M. yaluensis was close to B. springeri which is accordance with previously reported (Park et al. Citation2016 (Yang et al., Citation2016) ). The M. tungtingensis and M. fukiensis showed a closest phylogenetic relationship, then clustered with M. kiatingensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in National Center for Biotechnology Information(https://www.ncbi.nlm.nih.gov/)with accession number MN970213.
Additional information
Funding
References
- Chen DX, Chu WY, Liu XL, Nong XX, Li YL, Du SJ, Zhang JS. 2012. Phylogenetic studies of three sinipercid fishes (Perciformes: Sinipercidae) based on complete mitochondrial DNA sequences. Mitochondrial DNA. 23(2):70–76.
- Huang SP, Chen IS, Zhao Y, Shao KT. 2018. Description of a new species of the Gudgeon genus Microphysogobio Mori 1934 (Cypriniformes: Cyprinidae) from Guangdong Province, Southern China. Zool Stud. 57:e58.
- Huang SP, Zhao Y, Chen IS, Shao KT. 2017. A new species of Microphysogobio (Cypriniformes: Cyprinidae) from Guangxi Province, Southern China. Zool Stud. 56:e8.
- Park CE, Park G-S, Kim M-C, Kim K-H, Park HC, Lee I-J, Shin J-H. 2016. Mitochondrial genome of the Korean endemic species Microphysogobio yaluensis (Teleostei, Cypriniformes, Cyprinidae). Mitochondrial DNA A. 27(5):3557–3559.
- Yang X-G, Chen I-S, Ren Y-P, Shen C-N. 2016. The complete mitochondrial genome of Fujian rod gugeon Microphysogobio fukienensis (Nichols) (Cypriniformes, Cyprinidae). Mitochondrial DNA. 27(2):1473–1475. doi:10.3109/19401736.2014.953098.
- Zou YC, Xie BW, Qin CJ, Wang MY, Yuan DY, Li R, Wen ZY. 2017. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 39(7):767–778.
