Abstract
The genus Hiatella is one of most abundant and widespread marine bivalves. To date, its intra-generic phylogeny remains disputed and mitogenome information is therefore much needed. Here, we first report the complete circular mitogenome of Hiatella sp. J that is distributed in the coast of Asia Pacific. The total length of this mitochondrial genome is 21,233 base pairs. It consists of 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs, and a major noncoding region (MNR). Phylogenetic analysis of COI from 25 species (Hiatellidae) revealed that the Hiatella sp. J was closely related to Asian Hiatella in the family Hiatellidae. This Hiatella mitogenome provides new molecular data for the further taxonomic and phylogenetic studies of the genus Hiatella of marine bivalves.
Keywords:
The marine bivalve genus Hiatella is extremely abundant with high diversity in morphology and great plasticity in the mode of life history. As their shell shapes and modes of life history are weakly correlated with the suggested taxonomical identity, there is still no generally accepted intra-generic phylogeny exists at the moment (Hunter Citation1949; Coan et al. Citation2000). The Hiatella arctica has long been considered as a cosmopolitan species that comprises multiple lineages (Lubinsky Citation1980). Previous study of Laakkonen et al. (Citation2015) revealed that at least 13 of the recognized lineages represent distinct species. According to the phylogenetic evidence of mitochondrial COI, nuclear ANT and 28S rRNA genes, the Hiatella sp. J lineage in the east coast of Asian Pacific is a new Hiatella species (NCBI: txid1638755) (Laakkonen et al. Citation2015). To better understand the phylogenetic relationships of Hiatellidae, here we present the complete mitochondrial genome of Hiatella sp. J.
An individual sample of Hiatella sp. J was collected at the Laizhou Bay in Shandong province and deposited in our laboratory collection (voucher specimen number: OUC-MGB-2018-HO-01). The muscle tissue samples were collected, washed with 3.5× concentrated PBS, and preserved in liquid nitrogen. Total genomic DNA was extracted using a phenol-chloroform extraction protocol (Russell and Sambrook Citation2001). DNA sequencing was carried out using genomic DNA by an Illumina Xten platform. NOVOPlasty (Dierckxsens et al. Citation2017) and MITOS (Bernt et al. Citation2013) were utilized for mitogenome assembly and annotation, respectively. Maximum-likelihood (ML) was conducted on the partitioned matrices with FastTree (Price et al. Citation2009).
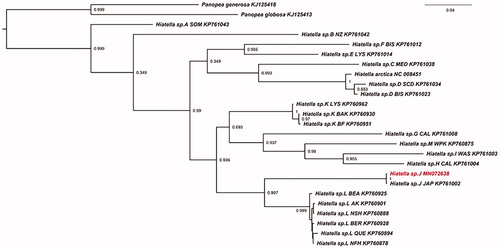
The complete circular mitogenome was 21,233 bp in length with 35.9% of GC content. It contained 13 protein-coding genes, 24 transfer RNAs, and 2 ribosomal RNAs. The mitogenomic sequence was submitted to GenBank with the accession number MN072638. Phylogenetic interference was performed with the COI genes of 22 Hiatella specimens (Laakkonen et al. Citation2015), outgrouped by two Panopea species. The ML phylogenetic tree depicted that our sample clustered with the Hiatella sp. J (), and had a close relationship with Hiatella sp. L that is distributed in Bering strait, Arctic ocean, Alaska, etc. This study provides a valuable mitogenome resource for better understanding of the molecular phylogeny of the genus Hiatella of marine bivalves.
Figure 1. The phylogenetic tree of COI based on ML. The accession number and sample sites for these species are as follows: Hiatella sp. L NFH (KP760878); Hiatella sp. L NSH (KP760888); Hiatella sp. L QUE (KP760894); Hiatella sp. L AK (KP760901); Hiatella sp. L BEA (KP760925); Hiatella sp. L BER (KP760928); Hiatella sp. J JAP (KP761002); Hiatella sp. M WPK (KP760875); Hiatella sp. K BAK (KP760930); Hiatella sp. K BF (KP760951); Hiatella sp. K LYS (KP760962); Hiatella sp. I WAS (KP761003); Hiatella sp. H CAL (KP761004); Hiatella sp. G CAL (KP761008); Hiatella sp. F BIS (KP761012); Hiatella sp. E LYS (KP761014); Hiatella sp. D BIS (KP761023); Hiatella sp. D SCD (KP761034); Hiatella sp. C MED (KP761038); Hiatella sp. B NZ (KP761042); Hiatella sp. A SOM (KP761043); Panopea generosa (KJ125418); Panopea globosa (KJ125413); Hiatella arctica (NC 008451).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
The data that support the findings of this study are openly available in GeneBank at [https://www.ncbi.nlm.nih.gov/nuccore/MN072638.1/], reference number [Accession number: MN072638].
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M and Stadler P F. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogen Evol. 69(2): 313–319.
- Coan E, Scott P, Bernard F. 2000. Bivalve seashells of western North America: marine bivalve mollusks from Baja California to Arctic Alaska. Stud Biodivers. Santa Barbara, California: Santa Barbara Museum of Natural History. 483–491.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(0e18.
- Hunter W. 1949. The structure and behavior of Hiatella gallicana (Lamarck) and H. arctica (L.), with special reference to the boring habit. Proc Sect B Biol. 3.63: 271–289.
- Laakkonen H, Strelkov P, Vainola R. 2015. Molecular lineage diversity and inter-oceanic biogeographical history in Hiatella (Mollusca, Bivalvia). Zool Scr. 44(4):383–402.
- Lubinsky I. 1980. Marine bivalve molluscs of the Canadian central and eastern Arctic: faunal composition and zoogeography. Ottawa: Department of Fisheries and Oceans Canada.
- Price M N, Dehal P S and Arkin A P. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol.26(7): 1641–1650.
- Russell D W and Sambrook J. 2001. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press. 1(6):1–28.
