Abstract
The mitochondrial genome of Phyllodiaptomus diaptomus was sequenced and assembled via Next-Generation Sequencing (NGS) and iterative assembly with a reference seed. The genome is 16446 bp long, A + T biased (69.4%), with 13 protein-coding genes, 22 tRNAs, and 2 rRNA. All protein-coding genes are initiated by a typical “ATN” codon. ND1, ND2, ATP6 genes are terminated with “TAG”, the other 10 genes are terminated with “TAA”. This is the first complete mitogenome published in the Diaptomidae. It provides molecular information useful to a better understanding of the phylogeny of calanoids.
Phyllodiaptomus tunguidus is a Chinese endemic and was first described by Shen and Tai (Citation1964) from the specimens collected from the Pearl River Delta in southern China. The original specimens of the species were stored in Zoological Museum, Institute of Zoology, Chinese Academy of Sciences (CAS). It belongs to the Calanoida, Diaptomidae and is mainly distributed in South China. P. tunguidus is a dominant species of zooplankton in most reservoirs of tropical and subtropical China (Lin et al Citation2003; Zhao and Han Citation2007). So far only one species of this genus has been found in China. Due to its large size and abundance, it has a strong effect on phytoplankton and it therefore plays an important role in tropical freshwater food webs. Complete mitochondrial genome data of such keystone species provide useful information for understanding its own phylogeny and the ecology of the order Calanoida (Jooste et al. Citation2019). Here, we sequence and annotate the mitochondrial genome of P. tunguidus and compare it to that of other copepods.
Living animals were collected from Liuxihe reservoir (113.46°E, 23.76°N) and fixed with absolute ethanol. The specimens (accession number COZOOP03003A) and extracted DNA were preserved at −20 °C in the Aquatic Collection of Institute of Hydrobiology, Jinan University, Guangzhou, China. We used the TIANamp Marine Animals DNA Kit (TIANGEN BIOTECH CO., LTD) to extract the genome. Genomic DNA was sequenced using a next-generation method. The assembly and annotation procedure followed Xu et al. (Citation2018) with the COI sequence of P. tunguidus as seed. The mitochondrial genome was annotated with the MITOS WebServer (Bernt et al. Citation2013) and verify via BLAST (Altschul et al. Citation1990). Transfer RNA genes were double-checked with tRNAscan-SE v.2 (Lowe and Eddy Citation1997) and ARWEN V.1.2.3 (Laslett and Canback Citation2008). A phylogenetic tree was constructed on 13 PCGs of 6 available mitogenomes of three Copepoda taxa from NCBI. Due to extensive gene reshuffling in this group (Minxiao et al. Citation2011) the sequences were separated into protein-coding genes, which were aligned separately via AliView (Larsson Citation2014). A Bayesian phylogenetic tree was constructed using a JTT substitution model in BEAST2 (Bouckaert et al. Citation2014). Finally, the synteny of all mitochondrial genes between P. tunguidus and Lovenula raynerae was analyzed using SimpleSynteny (Veltri et al. Citation2016).
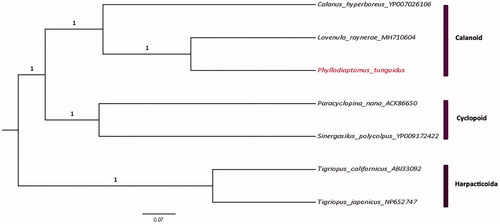
The total length of the P. tunguidus mitogenome is 16446 bp (GenBank accession number: MN927223, released on March 8, 2020), with A + T biased composition: A (34.6%), C (12.3%), G (18.2%), T (34.8%). Thirty-seven genes were annotated: 13 protein coding genes, 22 tRNA genes and 2 rRNA genes. All protein-coding genes are initiated by a typical “ATN” codon. COX1, COX2, COX3, ND2, ND3, ND4, ND5, ND6, ND4L, CYTB are end with TAA; ND2, ND1 and ATP6 end with TAG; tRNA genes have lengths ranging from 56 to 68bp, and they are folded into a typical cloverleaf structure. The Phylogenetic tree shows that copepods are split into three orders-level clades, namely Calanoida, Cyclopoida and Harpacticoida (). Phyllodiaptomus tunguidus is monophyletic with L. raynerae and the two calanoids have a sister taxon relationship with Calanus hyperboreus. Compared with the mitogenome of L. raynerae, a different arrangement of tRNAs was found, but no shift of PCGs was discovered.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MN927223.1, GenBank number: MN927223.1.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537.
- Jooste CM, Emami-Khoyi A, Gan HM, Wasserman RJ, Dalu T, Teske PR. 2019. The complete mitochondrial genome of Africa’s largest freshwater copepod, Lovenula raynerae. Mitochondr DNA Part B. 4(1):725–727.
- Larsson A. 2014. Aliview: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30(22):3276–3278.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Lin QQ, Duan SS, Hu R, Han B-P. 2003. Zooplankton distribution in trophic reservoirs, south China. Internat Rev Hydrobiol. 88(6):602–613.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Minxiao W, Sun S, Li CL, Shen X. 2011. Distinctive mitochondrial genome of Calanoid copepod Calanus sinicus with multiple large non-coding regions and reshuffled gene order: useful molecular markers for phylogenetic and population studies. BMC Genomics. 12:73.
- Shen J, Tai A. 1964. A description of eight new species in Calanoida in the the Pearl River Delta. Acta Zoological Sinica. 16(2):225–239.
- Veltri D, Wight MM, Crouch JA. 2016. SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 44(W1):W41–45.
- Xu SL, Guan ZY, Huang Q, Xu L, Vierstraete A, Dumont HJ, Lin QQ. 2018. The mitochondrial genome of Atrocalopteryx melli Ris, 1912 (Zygoptera: Calopterygidae) via lon Torrent PGM NGS sequencing. Mitochondrial DNA B. 3(1):115–117.
- Zhao SY, Han B-P. 2007. Structural analysis of zooplankton community in a large deep oligotrophic reservoir – Xinfengjiang Reservoir, South China. J. Lake Sci. 19(3):305–314.