Abstract
Liparis bootanensis is an epiphytic orchid distributed in tropical and subtropical regions of Asia, and has been listed as an endangered species in the Wildlife Conservation List. In this study, the complete chloroplast genome of L. bootanensis was assembled using Illumina sequencing data. The complete chloroplast (cp) genome is 158,325 bp in length, including a pair of invert repeats (IRA and IRB) regions of 26,700 bp, large single-copy (LSC) region of 86,584 bp, and small single-copy (SSC) region of 18,341 bp. The chloroplast genome contains 133 genes, including 83 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The phylogenetic analysis indicated that Oberonia japonica was closely related to L. bootanensis based on 17chloroplast genomes matrix of Orchidaceae.
The genus Liparis (Orchidaceae) comprises about 400 species and is widely distributed in the tropical and subtropical regions of Asia, Oceania and Americas, with only a few extending to the temperate zone (Chen et al. Citation2009; Lee et al. Citation2010; Lin and Yan Citation2013). Liparis species can be terrestrial, epiphytic or lithophytic (Chen et al. Citation2009). Morphologically, Liparis is characterized by resupinate flowers and curved column. Liparis bootanensis is an epiphytic orchid and widely distributed in tropical and subtropical regions of Asia. It usually grows on the tree trunk in forest and rocks or cliffs along valleys (Chen et al. Citation2009). It has been used in folk medicine in China (Liang et al. Citation2019). Due to the excessive collection and habitat destruction, the population size of L. bootanensis has declined during the last several decades. In this study, the complete chloroplast genome of L. bootanensis was assembled using Illumina sequencing data, which would be helpful for the formulation of conservation strategies and further research of L. bootanensis.
The total genomic DNA was extracted from fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The leaf sample of L. bootanensis was collected from Qinglong waterfall scenic area, Yongtai, Fujian province, China (25°46′N, 118°57′E). The voucher specimen is kept at the Herbarium of Fujian Agriculture and Forestry University (specimen code FAFU03805). Approximately 10 Gb of sequences data were extracted from the total sequencing output and input into Organelle PBA (Soorni et al. Citation2017) to assemble the chloroplast genome. Annotation of the chloroplast genome was performed using the Dual Organellar GenoMe Annotator (DOGMA) online tool (Wyman et al. Citation2004) and Geneious ver. 2019.1.1 (Li et al., Citation2019), then manually verified and corrected by comparison with L. loeselii (GenBank accession MF374688). Finally, we obtained a complete chloroplast genome of L. bootanensis and submitted to GenBank with an accession number (MN627759).
The total chloroplast genome sequence of L. bootanensisis 158,325 bp in length with GC content of 36.9%. It contains a pair of inverted repeats (IR) regions of 26,700 bp, a large single-copy (LSC) region of 86,584 bp, and a small single-copy (SSC) region of 18,341 bp.The chloroplast genome has a total of 133 genes, including 83 protein-coding genes, 38 tRNA genes, and 8 rRNA genes.
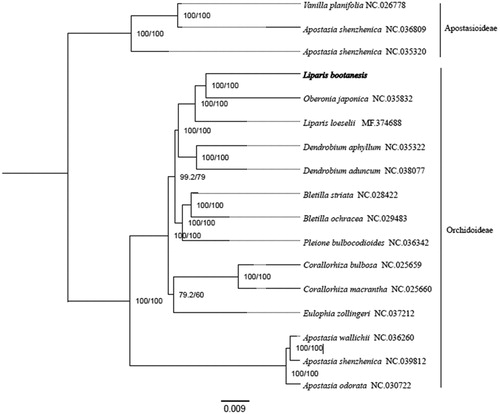
To explore the phylogenetic position of L. bootanensis, 16 complete chloroplast genomes of Orchidaceae (Apostasia odorata, A. shenzhenica, A. wallichii, Bletillastriata, B. ochracea, Pleione bulbocodioides, Corallorhizabulbosa, C. macrantha, Eulophia zollingeri, Dendrobium aphyllum, D. aduncum, L. loeselii, Oberonia japonica, Vanilla planifolia) were used to perform phylogenetic analysis. All the sequences were downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and the phylogenetic tree was constructed by RAxML (Stamatakis Citation2014). The results showed that O. japonica was most closely related to L. bootanensis ().
Disclosure statement
The author reports no conflict of interests. The author alone is responsible for the content and writing of the paper.
Data availability statements
Data openly available in a public repository that does not issue DOIs. The data that support the findings of this study are openly available in [National Center for Biotechnology Information] at [https://www.ncbi.nlm.nih.gov/], reference number [MN627759].
Additional information
Funding
References
- Chen SC, Wood JJ, Ormerod P. 2009. Liparis L.C. Richard. In: Wu ZY, Raven PH, Hong D, editors. Flora of China, vol. 25. St. Louis: Science Press, Beijing & Missouri Botanical Garden Press; p. 211–228.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee CS, Tsutsumi C, Yukawa T, Lee NS. 2010. Two new species of the genus Liparis (Orchidaceae) from Korea based on morphological and molecular data. J Plant Biol. 53(3):190–200.
- Li Y, Li Z, Hu Q, Zhai J, Liu Z, Wu S. 2019. Complete plastid genome of Apostasia shenzhenica (Orchidaceae). Mitochondrial DNA Part B. 4(1):1388–1389. doi:. 10.1080/23802359.2019.1591192
- Liang W, Guo X, Nagle DG, Zhang WD, Tian XH. 2019. Genus Liparis: are view of its traditional uses in China, phytochemistry and pharmacology. J Ethnopharmacol. 234:154–171.
- Lin L, Yan HF. 2013. A remarkable new species of Liparis (Orchidaceae) from China and its phylogenetic implications. PLoS One. 8(11):e78112.
- Soorni A, Haak D, Zaitlin D, Bombarely A. 2017. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genomics. 18(1):49.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysisand post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellargenomes with DOGMA. Bioinformatics. 20(17):3252–3255.