Abstract
Anemone flaccida has long-term been used in Chinese traditional medicine with the effects of anticancer, anti-inflammatory, antimicrobial properties, and immune regulation. However, the genomic information of this species is limited, which hinders its further medicinal application. In the present study, the complete chloroplast genome of A. flaccida was sequenced and assembled. The genome size was 157,614 bp in length, consisting of a pair of inverted repeat regions (IR, 31,184 bp), a large single copy (LSC, 79,055 bp), and a small single copy (SSC, 16,191 bp). A total of 138 genes were annotated, including 90 protein-coding genes, 40 tRNA genes, and eight rRNA genes. The GC content of the genome was 37.74%. A phylogenetic analysis on the basis of the whole chloroplast genome sequences further suggested a close relationship between A. flaccida, A. narcissiflora, and A. trullifolia. Collectively, the A. flaccida chloroplast genome provided new genomic resources which will improve its research and application in the future.
Anemone flaccida Fr. Schmidt is a perennial herb belonging to the Anemone genus in the Ranunculaceae family. The rhizomes of A. flaccida, known as ‘Di Wu’ in Chinese, is considered as a valuable Chinese traditional medicine for treatment of punch injury and rheumatoid arthritis (Liu et al. Citation2015). Previous studies on A. flaccida mainly focused on its chemical components, medicinal activities, and biosynthesis pathway about its main active ingredients (Han et al. Citation2016; Mo et al. Citation2019). However, the genomic resource of Anemone species was limited and the evolutionary relationship between A. flaccida and other Anemone species was not well investigated. In the present study, we sequenced and assembled the complete chloroplast (cp)genome of A. flaccida, and further investigated the phylogenetic relationship between A. flaccida and other representative Anemone species. All these will benefit both breeding and medicinal applications of A. flaccida in future.
The sample of A. flaccida used for sequencing was collected from Changyang County, Hubei province of China (30°17′N, 110°48′E), and the voucher specimen was deposited at the herbarium of Hubei University of Chinese Medicine (specimen code CY170410). The total genomic DNA was extracted from fresh leaves of the sample using a modified CTAB method (Doyle and Doyle Citation1987). A genomic library with an insert size of 500 bp was constructed and the library was sequenced using the Illumina Hiseq 4000 Sequencing System (Illumina, Hayward, CA) commercially in the Benagen Tech Solutions Company Limited (Wuhan, China). A total of 3.79 Gb of 150 bp paired-end reads were generated. The complete chloroplast genome sequence of A. flaccida was assembled using the program SPAdes 3.10.1 (Bankevich et al. Citation2012). Genome annotation was performed using the DOGMA (Wyman et al. Citation2004) and tRNAs were identified using the tRNAscan-SE v 1.21 (Schattner et al. Citation2005).
The complete chloroplast genome sequence of A. flaccida (GenBank accession number: MT317179) was 157,614 bp in length, containing a pair of inverted repeat (IR) regions of 31,184 bp each, separated by a large single-copy (LSC) region of 79,055 bp and a small single-copy (SSC) region of 16,191 bp. The overall GC content of the cp genome was 37.74%, and the corresponding values in LSC, SSC, and IR regions were 35.69, 31.88, and 41.87%, respectively. The circular genome contained 138 genes, including 90 protein-coding genes, 40 tRNA genes, and eight rRNA genes. Among these genes, 13 protein-coding genes, five tRNA genes, and five rRNA genes were duplicated in the IR region. With annotations of genes, 13 genes (atpF, ndhA, ndhB, rpl2, rpoC1, rps12, petB, petD, trnK-UUU, trnV-UAC, trnL-UAA, trnI-GAU, and trnA-UGC) were found to contain one intron, while two genes (clpP and ycf13) possessed two introns.
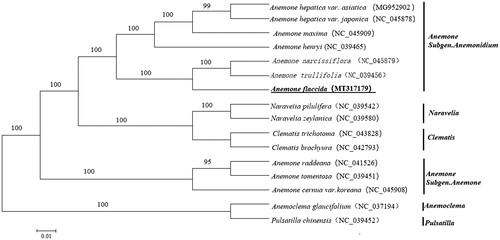
To investigate the evolutionary relationship of A. flaccida with other taxa within the tribe Anemoneae from the Ranunculaceae family, the whole chloroplast genome sequences of a panel of 16 plants, including 10 Anemone plants, two Naravelia plants, two Clematis plants and one Anemoclema plant and one Pulsatilla plant were used to construct a tree. Genome sequences were downloaded from the GenBank database and were aligned using MAFFT version 7 (Katoh and Standley Citation2013). We used MEGA 6 to generate a neighbour-joining tree (Tamura et al. Citation2013). As shown in the phylogenetic tree (), Anemone species from the subgenus Anemone and the subgenus Anemonidium were grouped into two clusters respectively, in which A. flaccida was closely related to both A. narcissiflora and A. trullifolia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI(National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MT317179.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Han L-T, Fang Y, Cao Y, Wu F-H, Liu E, Mo G-Y, Huang F. 2016. Triterpenoid saponin flaccidoside II from Anemone flaccida triggers apoptosis of nf1-associated malignant peripheral nerve sheath tumors via the mapk-ho-1 pathway. Onco Targets Ther. 9:1969–1979.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Liu Q, Zhu X-Z, Feng R-B, Liu Z, Wang G-Y, Guan X-F, Ou G-m, Li Y-L, Wang Y, Li M-M, et al. 2015. Crude triterpenoid saponins from Anemone flaccida (Di Wu) exert anti-arthritic effects on type II collagen-induced arthritis in rats. Chin Med. 10(1):20.
- Mo G-Y, Huang F, Fang Y, Han L-T, Pennerman KK, Bu L-J, Du X-W, Bennett JW, Yin G-H. 2019. Transcriptomic analysis in Anemone flaccida rhizomes reveals ancillary pathway for triterpene saponins biosynthesis and differential responsiveness to phytohormones. Chinese J Nat Med. 17(2):131–144.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan andsnoGPS web servers for the detection of tRNAs and snoRNAs. NucleicAcids Res. 33(Web Server):W686–W689.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of arganellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.