Abstract
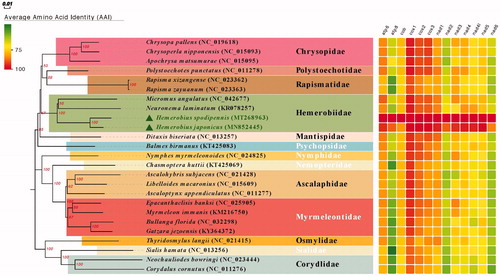
The complete mitochondrial genome of Hemerobius spodipennis Yang, Citation1987 was sequenced in this study. The complete mitochondrial genome is a typical double-stranded circular molecule of 16,343 bp (GenBank accession number: MT268963) comprising of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region. The gene order is identical to that of the putative ancestral arrangement of insects and other lacewings. All protein-coding genes initiate with ATN, except COI use CGA as start codons and terminate with TAG or TAA, expect ND5 and ND4 use TA– or a single T–– residue as the stop codon. All tRNAs, ranging from 63 to 72 bp, can be folded into typical clover-leaf secondary structure except for tRNASer(AGN), in which the dihydrouridine (DHU) arm did not form a stable stem-loop structure. The control region is 1433 bp long with an A + T content of 91.4%. In the sampled families of Neuroptera, each family showed a monophyletic cluster and Polystoechotidae + Rapismatidae, Hemerobiidae + (Chrysopidae + (Polystoechotidae + Rapismatidae)), are recovered in phylogenetic analyses with high supports.
Hemerobiidae, the brown lacewings, is the third largest family of Neuroptera, with about 650 species in the world and widely distributed. The genus Hemerobius is the largest genus of family Hemerobiidae, with about 250 species in the world (Oswald Citation1993, Citation2019; Monserrat Citation2000). In this study, we present the complete mitochondrial genome of the Chinese specie Hemerobius spodipennis Yang, Citation1987, which belongs to genus Hemerobius and important natural enemies because both their adults and larvae prey on aphid, scale insect, worm eggs, and mollusk insects (Yang Citation1981). The samples were collected in Purang County, Xizang, China (30°12′4″N, 81°15′10″E). Voucher specimen (No. HEME-00813) was deposited at the Entomological Lab of Nanjing Institute of Agricultural Sciences.
This mitochondrial genome is 16,343 bp long (GenBank accession number: MT268963). It includes the entire set of 37 genes (i.e. 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes) usually present in animal mitochondrial genomes and a control region. Gene order is identical to that of the putative ancestral arrangement of insects and other lacewings (Haruyama et al. Citation2011; Zhao et al. Citation2013; Cameron Citation2014; Zhang and Wang Citation2016; Zhao et al. Citation2016, Citation2020). There are a total of 43 overlapped nucleotides between genes in 16 locations, ranging from 1 to 7 bp in length; while there are totally 1694 bp intergenic nucleotides in 14 locations, ranging from 1 to 1433 bp in length.
ATN, GTG, TTG and GTT are accepted canonical mitochondrial start codons for invertebrate mtDNAs and most of the PCGs exhibit these start codons (Wolstrnholme Citation1992). Twelve protein-coding genes initiate with ATN as the start codon (ATG for COII, ATP6, COIII, ND4, ND4L and Cytb, ATT for ND2, ATP8, ND3, ND5, ND6, ATA for ND1). The exception is COI gene, which uses CGA as start codon. Conventional stop codons TAG and TAA are respectively distributed to two and nine protein-coding genes. However, ND5 terminates with TA–, and ND4 uses a single T–– residue as the stop codon.
There are 22 tRNA genes, ranging from 63 to 72 bp in length, and all of them can be folded into typical clover-leaf secondary structure expect for tRNASer(AGN), the dihydrouridine (DHU) arm of which forms a simple loop, as is common phenomenon in most insects. The length of lrRNA and srRNA is 1,324 bp and 784 bp, respectively. The A + T content of lrRNA and srRNA are determined to be 84.4% and 82.1%.
The control region is located between srRNA and tRNAIle and is 1433 bp in length with an A + T content of 91.4%, which is the most AT-rich region of this mitogenome. The A + T content of the whole genome, PCGs, tRNAs, and rRNAs was 80.2%, 78.2%, 79.3%, and 83.6%, respectively.
Phylogenetic relationship was inferred from phylogenetic analysis of the 13 protein-coding genes and generated by the neighbor-joining method (NJ) of MEGA7.0. Phylogenetic analyses showed the similar relationships among sampled families as shown in Winterton et al. (Citation2010). Each family showed a monophyletic cluster and the following clades were highly supported (): (1) Polystoechotidae + Rapismatidae; and (2) Hemerobiidae+(Chrysopidae+(Polystoechotidae + Rapismatidae)).
Acknowledgements
In particular, we thank Dr. Yunpeng Gai from Zhejiang University for his valuable contributions in technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT268963.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Haruyama N, Mochizuki A, Sato Y, Naka H, Nomura M. 2011. Complete mitochondrial genomes of two green lacewings, Chrysoperla nipponensis (Okamoto, 1914) and Apochrysa matsumurae Okamoto, 1912 (Neuroptera: Chrysopidae). Mol Biol Rep. 38(5):3367–3373.
- Monserrat VJ. 2000. New data on the Brown Lacewings from Asia (Neuroptera: Hemerobiidae). J Neuropterol. 3:61–97.
- Oswald JD. 1993. Revision and cladistic analysis of the world genera of the family Hemerobiidae (Insecta: Neuroptera). J New York Entomol S. 101:143–299.
- Oswald JD. 2019. Neuropterida species of the world. Version 3.0. [cited 2019 Nov 31]. http://lacewing.tamu.edu/Species-Catalogue/
- Winterton SL, Hardy NB, Wiegmann BM. 2010. On wings of lace: phylogeny and Bayesian divergence time estimates of Neuropterida (Insecta) based on morphological and molecular data. Syst Entomol. 35(3):349–378.
- Wolstrnholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yang JK. 1981. The brown lace-wings (Neuroptera: Hemerobiidae) of Wuyi Mountain. Wuyi Sci. 1:191–196.
- Yang JK. 1987. Agricultural insects, spiders, plant diseases and weeds of Xizang. Xizang: Xizang Renmin Press House. 1:463. pp.
- Zhang J, Wang XL. 2016. The complete mitochondrial genome of Myrmeleon immanis Walker, 1853 (Neuroptera: Myrmeleontidae) Mitochondrial DNA. 27(2):1439–1440.
- Zhao J, Li H, Winterton SL, Liu ZQ. 2013. Ancestral gene organization in the mitochondrial genome of Thyridosmylus langii (McLachlan, 1870) (Neuroptera: Osmylidae) and implications for lacewing evolution. PLOS One. 8(5):e62943.
- Zhao Y, Chen YJ, Zhao J, Liu ZQ. 2016. First complete mitochondrial genome from the brown lacewings (Neuroptera: Hemerobiidae). Mitochondrial DNA Part A. 27(4):2763–2764.
- Zhao Y, Shao HP, Zhang NN, Jing JQ. 2020. First complete mitochondrial genome of Hemerobius japonicus (Neuroptera: Hemerobiidae). Mitochondrial DNA Part B. 5(1):879–880.