Abstract
Muhlenbergia capillaris (Lam.) Trin. is an ornamental grass belonging to the genus Muhlenbergia Schreb. in the family Poaceae. To better understand its phylogenetic relationship with respect to the other species in the tribe Chloridoideae, the first complete chloroplast genome of Muhlenbergia was determined. The complete chloroplast genome of Muhlenbergia capillaris is 134,907 bp in length, consisting of one large single-copy (LSC) region of 80,175 bp, one small single-copy (SSC) region of 12,706 bp, and a pair of inverted repeat (IR) regions of 21,013 bp. The overall GC content of the genome is 38.1%. Further, maximum likelihood phylogenetic analysis with TVM + F+R3 model was conducted using 28 complete plastomes of the Poaceae, which support close relationships among species of Muhlenbergia, Hilaria Kunth, Distichlis Raf., and Bouteloua Lag., followed by those of Tragus Haller.
Muhlenbergia capillaris (Lam.) Trin., an ornamental grass distributed in Prairies, was assigned to the genus Muhlenbergia Schreb. in the family Poaceae. There are over 170 species in Muhlenbergia (http://www.plantsoftheworldonline.org/), and most of them are morphologically highly variable (Peterson et al. Citation1995). At the species level, the reported nuclear ITS and several chloroplast genomic markers were limited to resolve the phylogenetic and species identification problems in Muhlenbergia (Peterson et al. Citation2010). Here, we report the first complete chloroplast genome sequence of Muhlenbergia.
About 2 g leaf of M. capillaris in the nursery of Yunnan Forestry Technological College (Yunnan, China; Long. 102.771847°E, Lat. 25.097343°N, 1984 m) were collected for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Yunnan Forestry Technological College (Accession Number: HFTC-Qi Wang 002). The complete chloroplast genome was sequenced following Zhang et al. (Citation2016), and their 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. Clean paired-end reads were assembled with the GetOrganelle Kit (version 1.4.0) (Jin et al. Citation2018). The contigs were aligned using the publicly available chloroplast genome of Hilaria cenchroides Kunth (GenBank accession number KT168387) (Duvall et al. Citation2016) and annotated in Geneious 4.8.
The complete chloroplast genome of M. capillaris (SY001904), with a length of 134,907bp, was 73 bp and 1042 bp larger than that of H. cenchroides(134,824 bp, KT168387) and Bouteloua curtipendula (Michx.) Torr. (133,865 bp, KT168386) (Duvall et al. Citation2016). It was also 28 bp and 542 bp smaller than that of Tragus australianus S. T. Blake (134,935 bp, MK590077) and Distichlis bajaensis H. L. Bell (135,452 bp, KT168394) (Duvall et al. Citation2016; Anderson et al. Citation2019). The length of the large single-copy (LSC), inverted repeats (IRs), and small single-copy (SSC) regions of M. capillaris was 80,175 bp, 21,013 bp, and 12,706 bp, respectively. The overall G + C content was 38.1%.
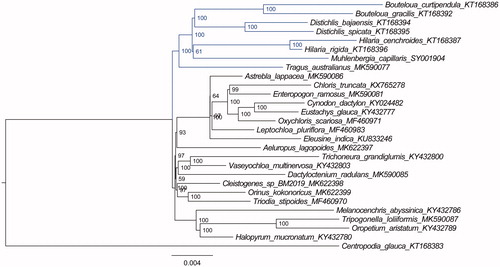
Based on 28chloroplast genome sequences, we reconstructed a phylogenetic tree () to confirm the relationship between M. capillaris and other related species with published chloroplast genomes in Chloridoideae, with Centropodia glauca (Nees) Cope as an outgroup. Maximum likelihood (ML) phylogenetic analyses were performed based on TVM + F+R3 model in the iqtree version 1.6.7 program with 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 59–100% bootstrap values at each node supported that close relationships among the species of Muhlenbergia, Hilaria Kunth, Distichlis Raf., and Bouteloua Lag., followed by Tragus australianus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data archiving statement
The chloroplast genome data of the Muhlenbergia capillaries will be submitted to Chloroplast Genome Database (https://lcgdb.wordpress.com). Accession numbers are SY001904.
Additional information
Funding
References
- Anderson B, Thiele K, Grierson P, Krauss S, Nevill PG, Small I, Zhong X, Barrett M. 2019. Recent range expansion in Australian hummock grasses (Triodia) inferred using genotyping-by-sequencing. AoB Plants. 11(2): 1–12.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722.
- Duvall MR, Fisher AE, Columbus JT, Ingram AL, Wysocki WP, Burke SV, Clark LG, Kelchner SA. 2016. Phylogenomics and plastome evolution of the chloridoid grasses (Chloridoideae: Poaceae). Int J Plant Sci. 177:1–12.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. https://doi.org/10.1101/256479
- Nguyen LT, Schmidt Ha v, Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Peterson PM, Romaschenko K, Johnson GP. 2010. A phylogeny and classification of the Muhlenbergiinae (Poaceae: Chloridoideae: Cynodonteae) based on plastid and nuclear DNA sequences. Am J Bot. 97(9):1532–1554.
- Peterson PM, Webster RD, Valdés-Reyna J. 1995. Subtribal classification of the New World Eragrostideae (Poaceae: Chloridoideae). SIDA. 16:529–544.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Sytem Evol. 54(3):219–227.