Abstract
The black carp (Mylopharyngodon piceus), native to eastern Asian, is a large, commercially valuable fish, and has been widely introduced to other countries. In this study, the mitochondrial gene sequence of gray black carp (M. piceus MT084757) in Foshan, Guangdong Province was first determined using the Sanger sequencing method. The mitochondrial DNA genome was 16,616 bp in length, including 13 protein-coding genes (PCGs), two rRNA genes, 22 transfer RNA genes, and a non-coding control region (D-loop). The overall nucleotide composition of the mitochondrial DNA is 32.04% A, 24.52% T, 15.68% C, 27.76% G, with 56.56% AT, respectively. Phylogenetic tree analysis suggests that the gray black carp (M. piceus MT 084757) is closely related to Elopichthys bambus and Squaliobarbus curriculus. The complete mitochondrial genome of the gray black carp (M. piceus MT 084757) would be useful for researching the causes of changes in body color.
The black carp (Mylopharyngodon piceus) is an important economic and ecological freshwater fish with a wide distribution range from the Pearl River to the Amur River, mainly in the plains south of the Yangtze River in China (Chen Citation1998; Xue-Ping Citation2016). Black carp (M. piceus), grass carp (Ctenopharyngodon idella), silver carp (Hypophthalmichthys molitrix), and bighead carp (Hypophthalmichthys nobilis) have contributed a lot to China’s aquaculture industry, so they are called “four famous farm fishes” in China (Li Citation1998). Black carp feeds on mollusks and are often used to control the overproduction of these organisms in freshwater aquaculture facilities (Ben-Ami and Heller Citation2001). It has been introduced to North America, Europe, Africa, and the Middle East (Haag Citation2008).
The gray black carp (M. piceus MT084757) was collected from Foshan, Guangdong (23°02’N, 113°06’E) in China, and the complete mitochondrial gene sequence was determined using the Sanger sequencing method. All samples (SHOUBC72) have been deposited in the College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China. Total genomic DNA was extracted from a single specimen (SHOUBC72_1) using the improved method with multi-well plates (Pall Corporation) (Yue and Orban Citation2005). Subsequently, based on the existing complete mitochondrial gene of the black carp (M. piceus NC_011141.1) (Wang et al. Citation2012), 29 pairs of primers were designed, the samples were amplified by PCR, and then sequenced using Sanger sequencing technology. The mitogenome was assembled using M. piceus NC_011141.1 as reference. The identification and location of protein-coding genes were determined by M. piceus NC_011141.1 too. The program tRNAscan-SE-2.0.5.tar.gz was used to predict the tRNA genes (http://lowelab.ucsc.edu/tRNAscan-SE/) (Chan and Lowe Citation2019). A physical map of the genome was generated using GenomeVX (Conant and Wolfe Citation2008). The analysis of other nucleotide/amino acid components and the construction of the evolutionary tree were using MEGA7 (Kumar et al. Citation2016).
The complete genome sequence of the gray black carp (M. piceus MT 084757) is 16,616 bp in size. The overall base composition was 32.04%A, 24.52%T, 15.68%C, 27.76%G, with 56.56% AT, respectively, containing 13 protein coding-genes (PCGs), two rRNA genes, 22 transfer RNAs genes, and a non-coding control region (D-loop). 11 PCGs started with ATG, but CO1 uses GTG and ND5 uses TTG as the start codon; Five PCGs were finished with TGA. The incomplete stop codon (TA–, T––) were found in seven genes (ND1, ND2, ND4L, ND5, ND6, ATP6, CO1, and CYTB). Gene overlap was observed at the border of eight genes (tRNAIle-tRNAGln, ATP6-ATP8, ND4-ND4L, and tRNAThr-tRNAPro). All 22 tRNAs distributed on the H and L strands were between 68 and 76 bp in length. 14 tRNA genes were encoded on the H and eight on the L strands. Most of tRNAs could form a common cloverleaf secondary structure, except tRNASer(AGY) gene without DHU stem (Sprinzl Citation1998; Chan and Lowe Citation2019). Two rRNA genes, 12SRNA, and 16SRNA were 964 bp and 1692 bp in size, respectively, which located between the tRNAPhe and tRNALeu(UUR) and separated by the tRNAVal gene. The control region was located between tRNAPhe and tRNAPro.
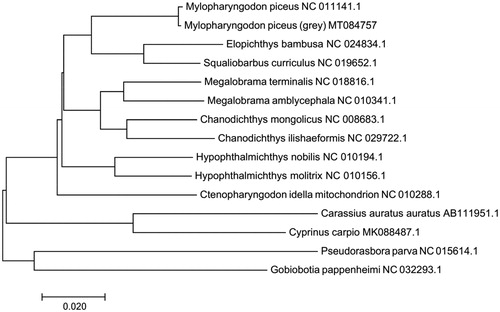
Based on the complete sequences of the mitochondrial genomes of 14 species, a phylogenetic tree was constructed using the neighbour-joining method (). The phylogenetic tree suggested that the gray black carp (M. piceus MT 084757) is closely related to Elopichthys bambus and Squaliobarbus curriculus.
Acknowledgments
We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Disclosure statement
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in the manuscript entitled, ‘Complete mitochondrial genome of gray black carp (Mylopharyngodon piceus).’
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT084757, reference numberMT084757.
Additional information
Funding
References
- Ben-Ami F, Heller J. 2001. Biological control of aquatic pest snails by the black carp Mylopharyngodon piceus. Biol Control. 22.2(2):131–138.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Chen YY. 1998. Fauna Sinica, Osteichthyes, Cypriniformes II. Beijing: Science Press; p. 1–16.
- Conant GC, Wolfe KH. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 24(6):861–862.
- Haag RW. 2008. Black carp: biological synopsis and risk assessment of an introduced fish. J North Am Benthol Soc. 27(3):800–802.
- Kumar S, Glen S, Koichiro T. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li SF. 1998. Genetic characterization of major freshwater culture fishes in China. Shanghai: Shanghai Scientific & Technical Publishers; p. 142–147.
- Sprinzl M. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26(1):148–153.
- Wang C, Wang J, Yang J, Lu G, Song X, Chen Q, Xu J, Yang Q, Li S. 2012. Complete mitogenome sequence of black carp (Mylopharyngodon piceus) and its use for molecular phylogeny of leuciscine fishes. Mol Biol Rep. 39(5):6337–6342.
- Xue-Ping Z. 2016. Establishment and characterization of a cell line derived from fin of Mylopharyngodon piceus. Freshwater Fish.
- Yue GH, Orban L. 2005. A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis. 26(16):3081–3083.