Abstract
We report the first mitochondrial genome sequences for the gray reef shark, Carcharhinus amblyrhynchos. Two specimens from the British Indian Ocean Territory were sequenced independently using two different next generation sequencing methods, namely short read sequencing on the Illumina HiSeq and long read sequencing on the Oxford Nanopore Technologies’ MinION sequencer. The two sequences are 99.9% identical and are 16,705 base pairs (bp) and 16,706 bp in length. The mitogenome contains 22 tRNA genes, two rRNA genes, 13 protein-coding genes and two non-coding regions; the control region and the origin of light-strand replication (OL).
Main text
The gray reef shark Carcharhinus amblyrhynchos, is a highly-social, reef-dependent species distributed widely in the tropical Indo-Pacific and currently listed as ‘Near Threatened’ in the IUCN Red List (Smale Citation2009). Populations have declined due to illegal fishing activities (Osgood and Baum Citation2015; Ferretti et al. Citation2018). Whilst there have been genetic studies conducted on the species (Holmes et al. Citation2009; Momigliano et al. Citation2015, Citation2017), its mitogenome has not yet been described.
We describe the complete mitochondrial genome of C. amblyrhynchos. Tissue was sampled as fin clips from two specimens in the British Indian Ocean Territory in March 2018. Specimen 1 (GenBank MT093205) was a female tagged at location −5.46386° 71.77841° and specimen 2 (GenBank MT104515) was a male tagged at −5.24956° 71.79906°. Samples were stored at Hopkins Marine Station before specimen 1’s tissue was transferred to Silwood Park, Imperial College London. The samples were then analyzed independently in the two laboratories. The DNA from specimen 1 is available at Silwood Park DNA & Tissue Bank (CITES GB038) under accession VS8956-20002085971; DNA from specimen 2 is available at Hopkins Marine Station, Stanford University under accession 020002232485. For specimen 1, genomic DNA was extracted using Qiagen’s Blood & Tissue Kit and was sequenced using an Illumina HiSeq. The mitochondrial genome sequence was assembled using ABySS v2.0.2 (Jackman et al. Citation2017) and GapCloser v1.12 (Luo et al. Citation2012). For specimen 2, the DNA was extracted and sequenced using the Oxford Nanopore Technologies’ MinION sequencer following Johri et al. (Johri et al. Citation2019). The MitoFish mitoannotator (Iwasaki et al. Citation2013) was used to annotate the sequences, and these were aligned against one another and mitogenomes from other Carcharhinid species using MUSCLE (Edgar Citation2004) within Geneious Prime (v2019.0.4). A phylogenetic tree was produced in Geneious Prime using MrBayes (Huelsenbeck and Ronquist Citation2001; Ronquist et al. Citation2012) plugin (v.3.2.6, substitution model: HKY85, burn-in length: 100,000) using the gray bamboo shark (Chiloscyllium griseum; NC_017882) and scalloped hammerhead shark (Sphyrna lewini; NC_022679) as outgroups.
The complete mitochondrial genomes are 16,705 bp (specimen 1) and 16,706 bp (specimen 2) in length. Each contains two rRNAs, 22 tRNAs, 13 protein-coding genes and a non-coding control region. The nucleotide base composition is identical with 31.5% A, 25.2% C, 13.2% G and 30.1% T, the overall GC content is 38.4%. The two sequences have 16,682 identical sites (99.9% pairwise identity). The differences include one base addition and 23 substitutions. Four substitutions result in a change to the amino acid sequence of COI. These differences could be due to the sequencing methods or represent evidence of population structure within the species in BIOT despite high spatial connectivity across the territory (Carlisle et al. Citation2019).
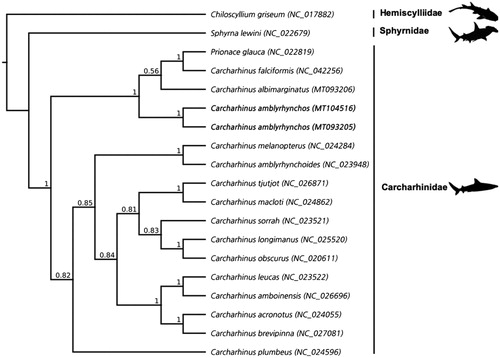
Whilst the fine-scale phylogenetic relationships within Carcharhinidae remain unresolved (Naylor et al. Citation2012), the tree () supports the placement of C. amblyrhynchos in a clade with C. albimarginatus, C. falciformis, and Prionace glauca. The low posterior probability that supports the placement of P. glauca with C. albimarginatus and C. falciformis suggests that further work is required to fully resolve the tree. However, the high support for deeper clades within Carcharhinus adds to calls for a taxonomic revision of P. glauca (Naylor et al. Citation2012; Johri et al. Citation2019). The new mitochondrial genomes presented here will aid in conservation genetics, environmental DNA and population studies as researchers move toward assessing populations using genome sequences.
Figure 1. Cladogram showing the phylogenetic relationship of species with complete mitogenome sequences in the genus Carcharhinus including Prionace glauca, with the scalloped hammerhead shark (Sphyrna lewini) and gray bamboo shark (Chiliscyllium griseum) as outgroups. The new sequences for the gray reef shark (Carcharhinus amblyrhynchos) are in bold. Families are indicated by vertical lines and represented by silhouettes accessed from PhyloPic (phylopic.org). Values at each node represent the Bayesian posterior probability at each node, GenBank accession numbers for each sequence are in brackets.
Author contributions
ND and SJ contributed to concept, bioinformatics and analysis, ND wrote the manuscript with comments from all authors, SJ contributed to sequencing and EAD provided laboratory support. BAB, VS and DC contributed to the concept of the manuscript and financial support of the project. BAB, TKC and DC contributed the samples.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT093205 reference number MT093205.1 and https://www.ncbi.nlm.nih.gov/nuccore/MT104515 reference number MT104515.1.
Additional information
Funding
References
- Carlisle AB, Tickler D, Dale JJ, Ferretti F, Curnick DJ, Chapple TK, Schallert RJ, Castleton M, Block BA. 2019. Estimating space use of mobile fishes in a large marine protected area with methodological considerations in acoustic array design. Front Mar Sci. 6:256. Available from: https://www.frontiersin.org/article/10.3389/fmars.2019.00256/full.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Ferretti F, Curnick D, Liu K, Romanov EV, Block BA. 2018. Shark baselines and the conservation role of remote coral reef ecosystems. Sci Adv. 4(3):eaaq0333.
- Holmes BH, Steinke D, Ward RD. 2009. Identification of shark and ray fins using DNA barcoding. Fish Res. 5(2-3):280–288.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, et al. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27(5):768–777.
- Johri S, Solanki J, Cantu VA, Fellows SR, Edwards RA, Moreno I, Vyas A, Dinsdale EA. 2019. ‘Genome skimming’ with the MinION hand-held sequencer identifies CITES-listed shark species in India’s exports market. Sci Rep. 9(1):1–13
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):2047–217X.
- Momigliano P, Harcourt R, Robbins WD, Jaiteh V, Mahardika GN, Sembiring A, Stow A. 2017. Genetic structure and signatures of selection in grey reef sharks (Carcharhinus amblyrhynchos). Heredity (Edinb). 119(3):142–153.
- Momigliano P, Harcourt R, Robbins WD, Stow A. 2015. Connectivity in grey reef sharks (Carcharhinus amblyrhynchos) determined using empirical and simulated genetic data. Sci Rep. 5(1):13229.
- Naylor GJ, Caira JN, Jensen K, Rosana K. a, Straube N, Lakner C. 2012. Elasmobranch phylogeny: a mitochondrial estimate based on 595 species. In: Biology of sharks and their relatives. 2nd (Carrier, J. C., Musick, J. A. & Heithaus, M. R.), 31–56. Boca Raton, FL: CRC Press
- Osgood GJ, Baum JK. 2015. Reef sharks: recent advances in ecological understanding to inform conservation. J Fish Biol. 87(6):1489–1523.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP . 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–42
- Smale M. 2009. Carcharhinus amblyrhynchos, Grey Reef Shark. IUCN Red List Threat Species. 8235:e.T39365A10216946.
