Abstract
Lactarius trivialis is a very common species and widely distributes in north temperate. In this study, a complete mitogenome of L. trivialis was assembled and annotated. The whole mitogenome of L. trivialis was a circular molecule with 42,366 bp in length, encoding 44 genes as follows: 19 coding genes, two rRNAs, and 23 tRNAs. The contents of four bases in mtDNA were A (39.65%), T (38.82%), C (11.30%), and G (21.53%), respectively. Phylogenetic analysis recovered that it is nested with other Lactarius spp. in the order Russulalles.
As a large genus in Agaricoid taxa, Lactarius Pers. (Russulaceae) has high biodiversity with more than 600 species and form ectomycorrhiza (Buyck et al. Citation2010, Citation2018). Lactarius spp. were divided into six subgenera based on morphological evidences (Buyck et al. Citation2018). However, molecular phylogenies did not support morphological classifications (Buyck et al. Citation2018). Additional molecular marks are needed for species phylogenetic analyses and evolutionary analyses. The target species, Lactarius trivialis, belongings to subgenus Piperites (Fr. ex J. Kickx f.) Kauffman, and it is a very common species with a wide distribution in north temperate. It can be easily identified by its characteristics in robust fruit bodies, zonate pileus without strigose and color change with aging, and smooth stipe (Lee et al. Citation2019). In this study, a complete mitochondrial genome of L. trivialis was reported, which will provide genomic data for phylogenetic and evolutionary investigations of Lactarius and Russulaceae.
The sample of L. trivialis was bought in Yunnan Mushuihua wild edible mushroom market in Kunming, China (25°0′N, 102°43’E). Total DNA was stored in Yunnan Agricultural University, Yunnan, China with voucher no. MG71. The raw data of L. trivialis (Mycobank: 201199) was retrieved from NCBI (SRR5803928, https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR5803928), then de novo assembled to a complete and circular mitogenome using GetOrganelle (Jin et al. Citation2018). Gene annotation was accomplished using The MFannot tool (Beck and Lang Citation2010) (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and manually checked in Geneious Primer 2020 (Biomatters, New Zealand). Sequences of Coding regions (CDS) and rRNAs were extracted, aligned, and concatenated using Geneious for phylogenomic analyses. Twenty species from Russulaceae, Boletales, and Agaricales were sampled, and Ganoderma lucidum (Curtis) P. Karst was selected as an outgroup. The best-fit model GTR + F+R5 using the Akaike information criterion (AIC) was applied to read the best model to construct Maximum-likelihood (ML) trees using IQ-TREE 1.6 (Lam-Tung et al. Citation2015).
The mitochondrial genome sequence of L. trivialis (GenBank accession MT267529) is a circular molecule with 42,366 bp in length, in which the contents of four bases were A (39.65%), T (38.82%), C (11.30%), and G (21.53%), respectively. The mitogenome encoded 44 genes as follows: 19 CDSs, two rRNAs, and 23 tRNAs. Typical ATG was used as the initiator codon and typical TAA as the terminator codon in all the 15 protein-coding genes (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and rps3). The gene order and organization of the L. trivialis are consistent with congener species in Lactarius (Li, Ren et al., Citation2019; Li, Wang et al., Citation2019).
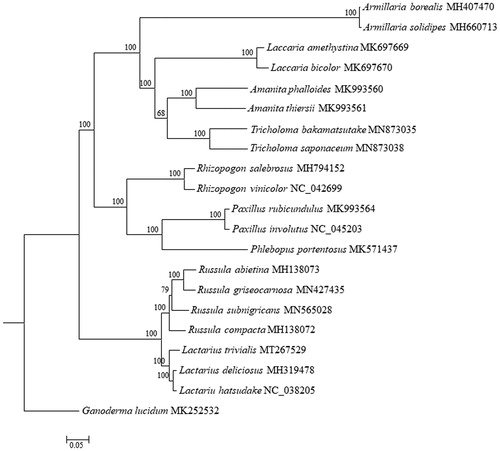
Phylogenomic analysis showed that L. trivialis clustered with other species of Lactarius spp. as a sister subclade to the subclade formed by Russula spp. (), both which belonged to Russulalles. This newly reported mitchondrional genome of L. trivialis provide important genomic information for species evolution and phylogenetic analyses of Lactarius and Russulaceae.
Figure 1. Phylogenetic analysis of Lactarius trivialis using maximum-likelihood of 15 protein-coding genes and two rRNA genes from 20 species from Russulaceae, Boletales, and Agaricales, and Ganoderma lucidum as outgroup. The bootstrap values were indicated close to the tree branches and each tip was followed by species name and accession number of GenBank.
Acknowledgments
The authors would like to thank Ms. Lu Tang and Dr. Yu Song for helps in software applications. We also give many thanks to professor Yang Dong from Yunnan Agriculture University for kindly permission to derive and analyze genome data of Lactarius trivialis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank with accession number MT267529. These data of Lactarius trivialis were derived from the following resources available in the public domain: NCBI Sequence Read Archive under accession number SRR5803928 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR5803928).
Additional information
Funding
References
- Beck N, Lang B. 2010. MFannot, organelle genome annotation websever. http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl.
- Buyck B, Hofstetter V, Eberhardt U, Verbeken A, Kauff F. 2018. Walking the thin line between Russula and Lactarius: the dilemma of Russula subsect. Ochricompactae. Fungal Diver. 89(1):240–267.
- Buyck B, Hofstetter V, Verbeken A, Walleyn R. 2010. Proposal 1919: to conserve Lactarius nom. Cons. (Basidiomycota) with a conserved type. Taxon. 59(1):295–508.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv 256479.
- Lam-Tung N, Schmidt HA, von Haeseler A, Bui Quang M. 2015. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Lee H, Wissitrassameewong K, Park MS, Verbeken A, Eimes J, Lim YW. 2019. Taxonomic revision of the genus Lactarius (Russulales, Basidiomycota) in Korea. Fungal Diver. 95(1):275–335.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the Subphylum Agaricomycotina. IJMS. 20(20):5167.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260.
