Abstract
The herb Isodon serra (Maximowicz) Kudô, which is widely distributed in China and its neighbor regions, is a well-known traditional Chinese medicinal plant. In this study, we characterized the complete plastid genome sequence of I. serra using Illumina sequencing data. The plastome is 152,676 bp in length and contains a typical quadripartite structure. The inverted repeat (IR), large-single copy (LSC) and small-single copy (SSC) regions each has 25,716 bp, 83,564 bp, and 17,680 bp. The genome contains 80 protein coding genes (PCGs), 30 transfer RNAs (tRNA), and four ribosomal RNAs (rRNA). The phylogenetic result indicates I. serra together with genera Ocimum and Lavandula formed tribe Ocimeae clade.
The genus Isodon (Schrad. ex Benth.) Spach (Lamiaceae) comprises ca. 100 species, and is widely distributed from tropical and subtropical Asia to southern Africa (Li and Hedge Citation1994; Yu et al. Citation2014). Many Isodon species are used in popular folk medicine in China, such as Isodon rubescens (Sun et al. Citation2006). The herb Isodon serra (Maximowicz) Kudô, widely distributed in China and its neighbor regions (Li and Hedge Citation1994), is a well-known traditional Chinese medicinal plant for its antibacterial, anti-inflammatory and antitumor activities (Huang et al. Citation2019). To our knowledge, the Isodon plastid genome has not been reported in the literature. Herein, we characterized the complete plastid genome sequence of I. serra using Illumina sequencing data. The plastid genome (GenBank accession no. MT317099) will be useful in the evolutionary and medicinal studies of Isodon.
Fresh leaves of an individual of I. serra was collected from Hutchison Whampoa Guangzhou Baiyunshan Chinese Medicine Co., Ltd (N23°11′11″, E113°15′57″), and used to extracted Genomic DNA using a modified CTAB protocol (Doyle and Doyle Citation1987). The voucher specimen is deposited at the Herbarium of South China Botanical Garden (Herbarium code: IBSC) with the accession number Zhy-Z3. Illumina paired-end library was constructed and sequencing based on Illumina Hiseq X Ten platform at Beijing Genomics Institute (Wuhan, China). The genomes were assembled using SPAdes v3.10.1 (Bankevich et al. Citation2012) and Geneious Prime 2019 (Biomatters, Ltd, Auckland, New Zealand) was subsequently used to close gaps. The coding and tRNA genes were annotated using Geneious Prime 2019. The maximum likelihood (ML) tree was reconstructed using RAxML (Stamatakis Citation2014) with the GTR + gamma model and 1000 bootstraps.
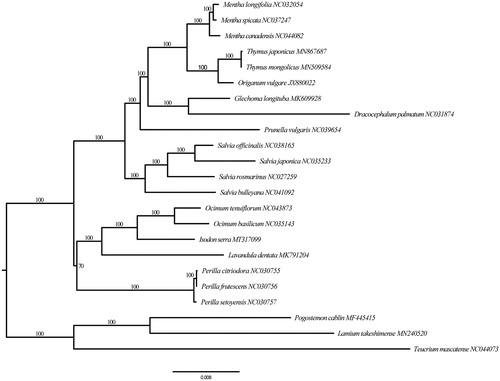
The plastome of I. serra is 152,676 bp in length and contains a typical quadripartite structure, comprising two copies of inverted repeat (IR) regions (25,716 bp), a large-single copy (LSC) region (83,564 bp), and a small-single copy (SSC) region (17,680 bp). It has 80 protein coding genes (PCGs), 30 transfer RNAs (tRNA), and four ribosomal RNAs (rRNA). The GC content of the genome is 37.6%, and varies among three regions. Specifically, LSC, SSC, and IR regions each has 35.7, 31.1, and 43.1%. The ML tree based on 80 PCGs shows that tribe Elsholtzieae (only represented by Perila in this study) is sister to the remainder of the subfamily Nepetoideae, followed by a sister group tribe Ocimeae + tribe Mentheae. I. serra together with genera Ocimum and Lavandula formed tribe Ocimeae clade (). This evolutionary relationships recovered here are consistent with the most recent analyses (Chen et al. Citation2016; Li et al. Citation2016).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number MT317099.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen YP, Drew BT, Li B, Soltis DE, Soltis PS, Xiang CL. 2016. Resolving the phylogenetic position of Ombrocharis (Lamiaceae), with reference to the molecular phylogeny of tribe Elsholtzieae. Taxon. 65(1):123–136.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang S, Hu W, Zeng S, Mo X, Wang Y. 2019. Comparative transcriptome analysis and simple sequence repeat marker development for two closely related Isodon species used as ‘Xihuangcao’herbs. Trop J Pharm Res. 18(1):75–84.
- Li B, Cantino PD, Olmstead RG, Bramley GL, Xiang CL, Ma ZH, Tan YH, Zhang DX. 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci Rep. 6(1):1–18.
- Li HW, Hedge IC. 1994. Lamiaceae. Flora of China. Beijing & St. Louis: Missouri Botanical Garden Press & Science Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun HD, Huang SX, Han QB. 2006. Diterpenoids from Isodon species and their biological activities. Nat Prod Rep. 23(5):673–698.
- Yu XQ, Maki M, Drew BT, Paton AJ, Li HW, Zhao JL, Conran JG, Li J. 2014. Phylogeny and historical biogeography of Isodon (Lamiaceae): rapid radiation in south-west China and Miocene overland dispersal into Africa. Mol Phylogenet Evol. 77:183–194.