Abstract
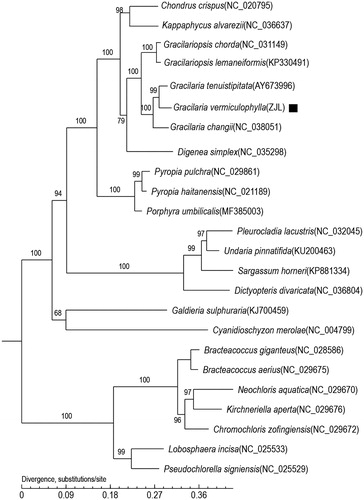
In this study, we sequenced and annotated the complete chloroplast genome of Gracilaria vermiculophylla (GenBank accession number: MN853882). The total length of the chloroplast genome of G. vermiculophylla is 180,262 bp, including 191 protein-encoding genes, 30 tRNA genes and 3 rRNA genes. The phylogenetic tree, which is based on core genes, shows that G. vermiculophylla clustered into the Gracilaria clade and has close genetic relationships with algae Gracilaria tenuistipitata and Gracilaria changii. These data will enable a better understanding of the phylogenetic status of G. vermiculophylla.
Gracilaria vermiculophylla belongs to the phylum Rhodophyta, class Florrdeophyceae, order Gracilariaceae, and family Gracilaria (Zeng et al. Citation2005). G. vermiculophylla was originally described in Japan and has a native distribution in the East of Asia. During the past three-to-four decades, this seaweed has successfully invaded many coastal habitats around the globe (Rueness Citation2005). Gracilaria vermiculophylla is one of the most important raw materials for the agar industry (Zhang et al. Citation2005) and is also a good heavy metal adsorption alga, which can significantly improve the water environment (Riosmena et al. Citation2010). With the recent development of genomics, an increasing number of chloroplast genomes of macroalgae have been sequenced and published (Leliaert and Lopez-Bautista et al. Citation2015; Melton et al., Citation2015; Cai et al. Citation2017); however, the chloroplast genome of G. vermiculophylla, an important economic alga, has not previously been reported. Therefore, we determined the complete chloroplast genome sequence of G. vermiculophylla.
Gracilaria vermiculophylla was collected from the Qinhunangdao coastal area (39°49′54.69″N, 119°31′30.50″E) and cultured in the laboratory with VSE media at 20 °C under a light intensity of 100 μmol−2 s−1 (Zhou et al. Citation2016a, Citation2016b). The specimens were preserved at the Marine Ecology Research Center of the First Institute Oceanography, Ministry of Natural Resources in Qingdao (Accession number: ZJL06).
The shape of the chloroplast genome of G. vermiculophylla is a double-stranded closed loop. The GenBank accession number is MN853882. The overall base composition of the G. vermiculophylla chloroplast genome is 25.99% A, 25.85% T, 24.08% C, and 24.08% G. The complete chloroplast genome is 180,262 bps long. There are 191 protein-coding genes in the genome, including 30 ycf genes, 27 rpl genes, 19 rps genes, 18 psb genes, 11 psa genes, 9 pet genes, 8 atp genes, 5 apc genes, 4 rpo genes, 3 acc, sec and cpc genes, 2 ccs, cpe, dna, ilv, inf, odp, rbc, suf, thi, and trp genes. In addition, there are some genes that are contained only once in the genome, such as acp, arg, bas, car, cbb, cem, chl, clp, dfr, dsb, fab, ftr, fts, glt, gro, lys, moe, nbl, ntc, omp, pbs, pgm, pre, rne, and rnz. In addition, the chloroplast genome contains 30 tRNA genes and 3 rRNA genes (rns, rrn5, and rnl), which are all non-coding. All the coding genes begin with ATG, except for infC, psaF, rbcS, and rps8, which begin with GTG. The termination codons for apcD, dsbD, odpB, petB, petL, psaC, psaD, psbA, psbJ, psbK, rpl27, rpl34, rps14, rps2, rps3, rps5, and trpG are TAG. However, acpP, apcD, atpF, lysR, petF, psaF, psbN, rpl33, rpl5, rpoC1, rps16, rps17, ycf29, ycf52, and ycf58 terminate with TGA. The remaining 159 genes end with TAA.
This is the first record of the chloroplast genome of G. vermiculophylla. The phylogenetic relationship of G. vermiculophylla with other species was estimated using a phylogenetic tree that was constructed based on the core genes of G. vermiculophylla as well as 24 other species. The phylogenetic tree () shows that G. vermiculophylla clustered into the Grateloupia clade and has close genetic relationships with algae Grateloupia filicina and Grateloupia taiwanensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov/ with the accession number is MN853882.
Additional information
Funding
References
- Cai C, Wang L, Zhou L, He P, Jiao B. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One. 12(9):e0184196.
- Leliaert F, Lopez-Bautista JM. 2015. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genom. 16(1):20.
- Melton JT, III, Leliaert F, Tronholm A, Lopez-Bautista JM. 2015. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp UNA00071828 (Ulvophyceae, Chlorophyta). PLoS One. 10(4):e0121020.
- Riosmena RR, Talavera SA, Acosta VB, Gardner S. 2010. Heavy metals dynamics in seaweeds and seagrasses in Bahía Magdalena, BCS, México. J Appl Phycol. 22(3):283–291.
- Rueness J. 2005. Life histories and molecular sequences of Gracilaria vermiculophylla. Phycologia. 44(1):120–128.
- Zeng CK, Wang SJ, Liu SJ. 2005. Seaweed Cultivation. Shanghai (China): Shanghai science and technology press; p. 225–250.
- Zhang HY, He PM, Chen CF, Dai YJ, Shi MJ. 2005. Effects of Porphyra yezoensis cultivation on inorganic nitrogen concentration in sea water. Environ Sci Technol. 28(4):44–45.
- Zhou LJ, Wang LK, Zhang JH, Cai CE, He PM. 2016a. Complete mitochondrial genome of Ulva linza, one of the causal species of green macroalgal blooms in Yellow Sea, China. Mitochondr DNA B. 1(1):31–33.
- Zhou LJ, Wang LK, Zhang JH, Cai CE, He PM. 2016b. Complete mitochondrial genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondr DNA B. 1(1):76–78.