Abstract
Paris delavayi Franchet is a perennial herb of the family Melanthiaceae. In this study, the complete chloroplast (cp) genome sequence of P. delavayi was characterized. The cp genome is 164,195 bp in length and contains a pair of inverted repeats (33,415 bp) separated by a large (84,400 bp) and small (12,965 bp) single-copy regions. A total of 112 unique genes were predicted, including 78 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The phylogenetic analysis suggested that P. delavayi is sister to P. mairei but with low support.
Paris delavayi Franchet is a perennial herb of the family Melanthiaceae and mainly distributed in Yunnan, Guizhou, Hubei and Hunan provinces of China (Li Citation1984). In Chinese Pharmacopeia, two other Paris plants: P. polyphylla var. yunnanensis and P. polyphylla var. chinensis are recorded as the original plants of Chonglou but become rare and endangered due to aggressive harvesting in recent decades. Many studies have reported that the major bioactive saponins have been also isolated from the rhizomes of P. delavayi (Liu et al. Citation2006; Fu et al. Citation2012; Liu et al. Citation2016; Huang et al. Citation2017). Therefore, P. delavayi could be further utilized as the potential medical resource. However, little genetic information is known about this species. To better understand and utilize this species, we sequenced and analyzed the complete chloroplast (cp) genome of P. delavayi using high-throughput sequencing technology.
The specimen (lpssy0305) was collected from Yushe National Forest Park (Liupanshui, Guizhou, China; 104°48′E, 26°27′N, 2,235 m) and deposited at the herbarium of the Liupanshui Normal University (LPSNU). Genomic DNA was extracted from the fresh leaves as previously described (Zhang et al. Citation2019) and used for the library construction and Illumina sequencing. Approximately 6 Gb raw data were used for the cp genome assembly using SPAdes (Bankevich et al. Citation2012). The cp genome annotation was accomplished using PGA (Qu et al. Citation2019).
The complete cp genome of P. delavayi (accession number MT038210) is 164,195 bp in length with a typical quadripartite structure containing two inverted repeats (IRs) of 33,415 bp, a large single copy (LSC) region of 84,400 bp and a small single copy (SSC) region of 12,965 bp. The overall GC content of the cp genome is 37%. A total of 112 unique genes consist of 78 protein-coding genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes, which is little different from other species of Paris (Huang et al. Citation2016). Among these genes, 14 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain one intron and three genes (clpP, rps12 and ycf3) have two introns.
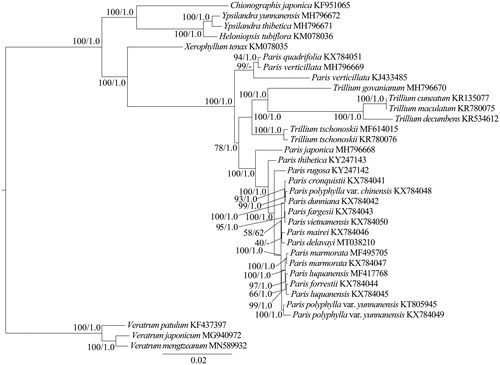
The genus Paris comprises about 27 species accepted in The Plant List (http://www.theplantlist.org/) and divides into two subgenera, Paris and Daiswa (Huang et al. Citation2016). In recent years, several new Paris species, such as P. lihengiana (Xu et al. Citation2019), P. tengchongensis (Ji et al. Citation2017) and P. nitida (Wang et al. Citation2017) were reported. In this study, we constructed the phylogenetic tree and analyzed the phylogenetic position of P. delavayi based on the maximum likelihood (ML) and Bayesian inference (BI) methods (Ronquist et al. Citation2012; Stamatakis Citation2014). Thirteen species (Veratrum patulum, V. japonicum, V. mengtzeanum, Chionographis japonica, Ypsilandra yunnanensis, Y. thibetica, Heloniopsis tubiflora, Xerophyllum tenax, Trillium govanianum, T. cuneatum, T. maculatum, T. decumbens and T. tschonoskii) from other six genera of Melanthiaceae were used as the outgroups. The cp genomes of P. delavayi and previously published species of Paris were used for phylogenetic analysis. The phylogenetic tree () illustrates that P. delavayi is sister to P. mairei but with weak support. Therefore, the phylogenetic position of P. delavayi will be further studied in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/, reference number MT038210.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Fu S, Li N, Liu Z, Gao W, Man S. 2012. Determination of seven kinds of steroidal saponins in plants of Paris L. from different habitats by HPLC. Chin Trad Herb Drugs. 43:2435–2437.
- Huang Y, Li X, Yang Z, Yang C, Yang J, Ji Y. 2016. Analysis of complete chloroplast genome sequences improves phylogenetic resolution in Paris (Melanthiaceae). Front Plant Sci. 7:1797–1808.
- Huang YY, Liu DH, Peng HS, Hao QX, Chen ML, Zhang AZ, Chen M, Kang LP, Huang LQ. 2017. Determination of eight steroidal saponins in 15 kinds of genus Paris. China J Chin Mater Med. 42(18):3443–3451.
- Ji Y, Yang C, Huang Y. 2017. A new species of Paris sect. Axiparis (Melanthiaceae) from Yunnan, China. Phytotaxa. 306(3):234–236.
- Li H. 1984. The phylogeny of the genus Paris L. Acta Bot Yunnanica. 6:351–362.
- Liu H, Huang Y, Zhang T, Wang Q, Chen X. 2006. Studies on chemical constituents of Paris delavayi Franchet. J China Pharm Univ. 37:409–412.
- Liu Y, Tian X, Hua D, Cheng G, Wang K, Zhang L, Tang H, Wang M. 2016. New steroidal saponins from the rhizomes of Paris delavayi and their cytotoxicity. Fitoterapia. 111:130–137.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang Z, Cai XZ, Zhong ZX, Xu Z, Wei N, Xie JF, Hu GW, Wang QF. 2017. Paris nitida (Melanthiaceae), a new species from Hubei and Hunan, China. Phytotaxa. 314(1):145–149.
- Xu Z, Wei N, Tan Y, Peng S, Ngumbau VM, Hu GW, Wang QF. 2019. Paris lihengiana (Melanthiaceae: Parideae), a new species from Yunnan, China. Phytotaxa. 392(1):45–53.
- Zhang SD, Zhang C, Ling LZ. 2019. The complete chloroplast genome of Rosa berberifolia. Mitochondrial DNA B. 4(1):1741–1742.