Abstract
Ficus erecta is a wild relative of F. carica, which is an important economic plant. Here, we determined the complete plastid genome of F. erecta using the Illumina paired reads to provide genomic feature resources. The whole plastid genome of F. erecta is 160,603 bp in length, containing two inverted repeats (IRs) of 25,899 bp separated by a large single-copy (LSC 88,640 bp) region and a small single-copy (SSC 20,165 bp) region. The complete plastome sequence of Ficus erecta will provide a useful resource for the evolutionary biology study as well as for the phylogenetic studies.
The genus Ficus includes about 735 species with a selection of syconiumsis, an extraordinary genus to investigate symbiotic relationships between flowers and their pollinators (Berg and Corner Citation2005, Galil Citation1973). Figs are very important economic plants as food and medicinal resources. Besides, figs also play important roles in ecology system in some regions. Ficus carica is one of the oldest cultivated plant for food resources but always susceptible to some fungus diseases such as Ceratocystis canker which is transmitted through soil (Kajitani and Masuya Citation2011). Ficus erecta is a wild relative of F. carica with resistance to Ceratocystis canker, provide a feasible way to combat soil-transmitted diseases. The genome of male F. erecta was assembled with high quality and provided insights into Ceratocystis canker resistance breeding strategies and the mechanisms of F. erecta resistance to diseases.
In this study, we downloaded total 3.9 G data of high-quality reads (NCBI accession # DRR187744) using the fastqdump software, which was released by the F. erecta genome sequencing project (Shirasawa et al. Citation2019). The individual for sequencing was a male F. erecta tree “FE-Hiroshima-1” which was grown naturally in Higashi-Hiroshima city in Japan. The sample was stored at Kazusa DNA Research Institute herbarium, Japan.
Sequenced reads were then used to assemble the complete plastid genome using NOVOPlasty with a rpoA CDS (NC_028185) as a seed sequence (Dierckxsens et al. Citation2017]). We performed the annotation using Geneious 10.2.2 and adjusted the protein-coding genes manually in order to make sure all of these genes were maintained as open reading frames. IR boundaries for the draft plastome were confirmed by BLAST. The physical map of the plastid genome was conducted via OGDraw online (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013). Finally, we obtained a chloroplast genome of F. erecta and submitted the whole genome to GenBank (MT093220).
The plastome of F. erecta is a double-stranded circular DNA of 160,603 bp in length with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 25,899 bp separated by a large single-copy (LSC) region and a small single-copy (SSC)region of 88,640 and 20,165 bp, respectively. The plastid genome of F. erecta contains 112 genes, including 78 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. The overall GC content of F. erecta plastid genome is 35.9% and the corresponding values in LSC, SSC, and IR regions are 33.5, 28.9, and 42.6%, respectively.
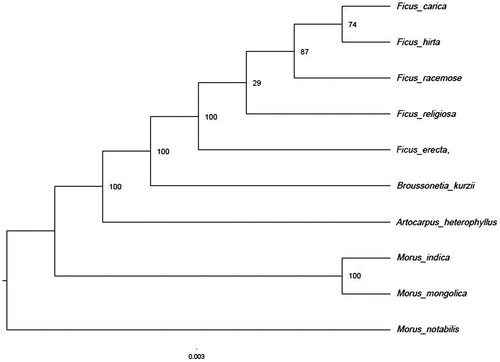
We also used the protein-coding genes of F. erecta and another 9 Moraceae species to reconstruct a maximum likelihood tree through RaxML (Stamatakis Citation2014) under the GTRGAMMA substitution model, with 1000 bootstraps on CIPRESwebsite (Miller et al. Citation2010). The result shows that Ficus formed the clade corresponding to Ficeae. (). The Moreae comprised a paraphyletic grade which was consistent with the previous study (Datwyler and Weiblen Citation2004).
Figure 1. Phylogenetic tree (maximum likelihood) based on the protein-coding genes of 10 species. Numbers above branch aremaximum parsimony bootstrap percentages.These accession numbers are as follows:Morusindica (DQ226511), Morusmongolica (KM491711), Morusnotabilis (MK211167), Ficus carica (NC_035237), Ficus hirta (MN364706), Ficus racemose (NC_028185), Ficus religiosa (NC_033979), Broussonetiakurzii (NC_041637), Artocarpusheterophyllus (MK303549), Antiaristoxicaria (NC_042884).
Acknowledgement
We are grateful to Kazusa DNA Research Institute, Japan, Institute of Fruit Tree and Tea Science, Japan, Hiroshima Prefectural Technology Research Institute, Agricultural Technology Research Center, Japan, Fukuoka Agriculture and Forestry Research Center, Japan, and National Institute of Genetics, Japan for their support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the results of this study are openly available in GeneBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MT093220 after the paper has been accepted. These raw data used in this study were derived from the following resources available in DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA).
References
- Berg CC, Corner E. 2005. Moraceae – Ficus. Flora Malesiana, Ser. I, 17/2. National Herbarium of the Netherlands. Leiden.
- Datwyler SL, Weiblen GD. 2004. On the origin of the fig: phylogenetic relationships of Moraceae from ndhF sequences[J]. Am J Bot. 91(5):767–777.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data[J]. Nucleic Acids Res. 45(4):18.
- Galil J. 1973. Pollination in dioecious figs: pollination of Ficus fistulosa by Ceratosolen hewitti. – Gdns. Bull Sing. 26:303–311.
- Kajitani Y, Masuya H. 2011. Ceratocystis ficicola sp. nov., a causal fungus of fig cankerin Japan. Mycoscience. 52(5):349–353.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), Orleans, 1–8.
- Shirasawa K, Yakushiji H, Nishimura R. 2019. The Ficus erecta genome aids Ceratocystis canker resistance breeding in common fig (F. carica)[J]. The Plant Journal. 102(6):1313-1322.[.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics. 30(9):1312–1313.
