Abstract
We report the complete mitochondrial genome for the Eospalax rufescens, a typical subterranean rodent species and endemic in China. The resulting E. rufescens mitogenome is 16,355 bases in size, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 noncoding control region (D-loop). The base compositions present highly biased toward A + T nucleotides. Moreover, twelve of all 13 PCGs initiated with ATN start codon, whereas ND1 began with GTG start codon. Stop codons in 13 PCGs were all typical types except incomplete stop codon T for ND4. We further provide a Maximum Likelihood phylogenetic tree showing relationships among E. rufescens and other common subterranean rodents in family Spalacidae.
The Qinling zokor, Eospalax rufescens, is a typical subterranean rodent species and endemic in China (Wang Citation2003) belonging to subfamily Myospalacinae, family Spalacidae (Norris et al. Citation2004). Here, we provide the first complete mitogenome of E. rufescens (GenBank accession: MH933740).
The samples of E. rufescens was collected from Shaanxi province of China (N34.13°, E107.15°, altitude 1783 m) and currently stored at Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences under voucher number TBX-03. Total genomic DNA was extracted from muscle using DNeasy Tissue Kit (QIAGEN). An Illumina library was generated from genomic DNA and sequenced on an Illumina Hiseq 2500 platform. The mitochondrial genomes were reconstructed by MitoZ (Meng et al. Citation2019) with the default parameters, and the complete mitogenome sequence of Myospalax aspalax (GenBank accession number: KP724691) was used as a reference. The Maximum Likelihood phylogenetic tree was conducted with RA × ML v8.2.12 (Stamatakis Citation2014) through the online CIPRES Science gateway (Miller et al. Citation2010).
A total of 35,172,146 sequence reads were generated by next-generation sequencing after removing adaptor polluted reads, more than 5% Ns reads and low-quality sequences. The accurate annotated mitochondrial genome sequence of E. rufescens is a circular double-strand DNA molecule of 16,350 bp, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and 1 non-coding region. The arrangement of the multiple genes is similar in line with other Myospalacinae species (Liu et al. Citation2011; Su et al. Citation2013; Li et al. Citation2016; Yuan et al. Citation2016; Cai et al. Citation2019). The overall base composition of the heavy strand is 32.81% A, 23.80% C, 31.12% T, and 12.96% G. The base compositions present highly biased toward A + T nucleotides. The heavy strand (H-strand) encodes 2 rRNA genes, 12 PCGs and 14 tRNA genes, the nicotinamide adenine dinucleotide dehydrogenase subunit 6 (ND6) gene and 8 other tRNA genes are encoded on the L-strand.
Nine of all 13 PCGs initiated with an ATG start codon except for ND1, ND2, ND3, and ND5, which began with GTG, ATT, ATA, and ATT start codons, respectively. Eight of the 13 PCGs use TAA as stop codon. The ND1, ND2, CO1 and ND6 are stopped with TAG, and ND4 end with incomplete stop codon T. The tRNA genes range from 60 to 75 bp in length and employ the anticodons typical of vertebrate mt-tRNAs. All the tRNAs had a typical secondary structure (cloverleaf structure) except the tRNA-Ser (GCT), whose complete dihydrouridine arm was lacking. The 12S and 16S ribosomal RNA genes are 939 and 1571 bp long, respectively. In our study, the noncoding control region of the E. rufescens mtDNA is 937 bp long.
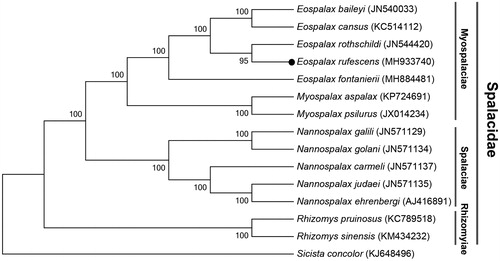
The results of phylogenetic analysis displayed that E. rufescens formed a clade with other species in subfamily Myospalacinae, and Rhizomyinae was a basal clade relative to others within Spalacidae (). This mitogenome sequence of E. rufescens would help in evolutionary biology study of the Mysopalacinae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The data that support the findings of this study are openly available in NCBI Sequence Read Archive (SRA) at https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=search_obj, reference number SRR11637900.
Additional information
Funding
References
- Cai Z, Zhang Y, Gao H, Song P, Zhang J, Zhang T. 2019. The complete mitochondrial genome of Chinese zokor (Eospalax fontanierii). Mitochondr DNA B. 4(1):153–154.
- Liu Z, Li Y, Shi F, Lu J, Li M, Wang Z. 2011. Mitochondrial genome of Plateau zokor Myospalax baileyi. Mitochondrial DNA. 22(5–6):174–175.
- Li Y, Lu J, Wang Z. 2016. Complete mitochondrial genome of Manchurian zokor (myospalax psilurus). Mitochondr DNA A DNA Mapp Seq Anal. 27(2):1461–1462.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Miller MA, Pfeiffer WT, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, in Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA pp 1 - 8.
- Norris RW, Zhou K, Zhou C, Yang G, William KC, Honeycutt RL. 2004. The phylogenetic position of the zokors (myospalacinae) and comments on the families of muroids (rodentia). Mol Phylogenet Evol. 31(3):972–978.
- Su J, Wang J, Hua L, Gleeson D, Ji W. 2013. Complete mitochondrial genome of the Gansu zokor, Eospalax cansus (rodentia, spalacidae). Mitochondr DNA. 24(6):651–653.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang Y. 2003. A complete checklist of mammal species and subspecies in China. A taxonomic and geographic reference. Beijing: China Forestry Publishing House (Chinese).
- Yuan S, Lu Z, Wu X, Fu H, Bao D, Malqin H, Yang S. 2016. Complete mitochondrial genome of Myospalax aspalax (rodentia, spalacidae). Mitochondr DNA A DNA Mapp Seq Anal. 27(6):4250–4251.